Abstract
Key message
The elite ZmCCT haplotypes which have no transposable element in the promoter could enhance maize resistance to Gibberella stalk rot and improve yield-related traits, while having no or mild impact on flowering time. Therefore, they are expected to have great value in future maize breeding programs.
Abstract
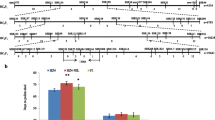
A CCT domain-containing gene, ZmCCT, is involved in both photoperiod response and stalk rot resistance in maize. At least 15 haplotypes are present at the ZmCCT locus in maize germplasm, whereas only three of them are found in Chinese commercial maize hybrids. Here, we evaluated ZmCCT haplotypes for their potential application in corn breeding. Nine resistant ZmCCT haplotypes that have no CACTA-like transposable element in the promoter were introduced into seven elite maize inbred lines by marker-assisted backcrossing. The resultant 63 converted lines had 0.7-5.1 Mb of resistant ZmCCT donor segments with over 90% recovery rates. All converted lines tested exhibited enhanced resistance to maize stalk rot but varied in photoperiod sensitivity. There was a close correlation between the hybrids and their parental lines with respect to both resistance performance and photoperiod sensitivity. Furthermore, in a given hybrid A5302/83B28, resistant ZmCCT haplotype could largely improve yield-related traits, such as ear length and 100-kernel weight, resulting in enhanced grain yield. Of nine resistant ZmCCT haplotypes, haplotype H5 exhibited excellent performance for both flowering time and stalk rot resistance and is thus expected to have potential value in future maize breeding programs.






Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733
Belcher A, Zwonitzer J, Cruz J, Krakowsky M, Chung C, Nelson R, Arellano C, Balint-Kurti P (2012) Analysis of quantitative disease resistance to southern leaf blight and of multiple disease resistance in maize using near-isogenic lines. Theor Appl Genet 124:433–445
Campoli C, Drosse B, Searle I, Coupland G, von Korff M (2012) Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J 69:868–880
Clements M, Kleinschmidt C, Maragos C, Pataky J, White D (2003) Evaluation of inoculation techniques for Fusarium ear rot and fumonisin contamination of corn. Plant Dis 87:147–153
Endo-Higashi N, Izawa T (2011) Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol 52:1083–1094
Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Garcia EW, Lebruska LL, Laurent M, Shen R, Barker D (2006) [3] illumina universal bead arrays. Methods Enzymol 410:57–73
Gao H, Jin M, Zheng X-M, Chen J, Yuan D, Xin Y, Wang M, Huang D, Zhang Z, Zhou K (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Nat Acad Sci 111:16337–16342
González-Schain ND, Díaz-Mendoza M, Żurczak M, Suárez-López P (2012) Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J 70:678–690
Grau C, Radke V, Gillespie F (1982) Resistance of soybean cultivars to Sclerotinia sclerotiorum. Plant Dis 66:506–508
Holland JB (2001) Epistasis and plant breeding. Plant Breed Rev 21:27–92
Hung H-Y, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF (2012) ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Nat Acad Sci 109:1913–1921
Kim S-K, Yun C-H, Lee JH, Jang YH, Park H-Y, Kim J-K (2008) OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228:355–365
Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63:18–30
Lisch D (2013) How important are transposons for plant evolution? Nat Rev Genet 14:49–61
Liu H, Liu H, Zhou L, Zhang Z, Zhang X, Wang M, Li H, Lin Z (2015) Parallel domestication of the heading date 1 gene in cereals. Mol Biol Evol 32:2726–2737
Liu H, Dong S, Sun D, Liu W, Gu F, Liu Y, Guo T, Wang H, Wang J, Chen Z (2016a) CONSTANS-Like 9 (OsCOL9) Interacts with receptor for activated C-kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS One 11(11):e0166249
Liu J, Shen J, Xu Y, Li X, Xiao J, Xiong L (2016b) Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot 67:5785–5798
Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran L-SP, Qin F (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6:8326
Miller TA, Muslin EH, Dorweiler JE (2008) A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227:1377–1388
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Nat Acad Sci 104:11376–11381
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh C-T, Emrich SJ, Jia Y, Kalyanaraman A, Hsia A-P, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia J-M, Deragon J-M, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Sibout R, Plantegenet S, Hardtke CS (2008) Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol 18:458–463
Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285
Stearns FW (2010) One hundred years of pleiotropy: a retrospective. Genetics 186:767–773
Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43:1160–1163
Tan J, Jin M, Wang J, Wu F, Sheng P, Cheng Z, Wang J, Zheng X, Chen L, Wang M, Zhu S, Guo X, Zhang X, Liu X, Wang C, Wang H, Wu C, Wan J (2016) OsCOL10, a CONSTANS-Like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol 57(4):798–812
Tao Y, Liu Q, Wang H, Zhang Y, Huang X, Wang B, Lai J, Ye J, Liu B, Xu M (2013) Identification and fine-mapping of a QTL, qMrdd1, that confers recessive resistance to maize rough dwarf disease. BMC Plant Biol 13(1):145
Tenaillon MI, Hollister JD, Gaut BS (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15:471–478
Thakur R, Leonard K, Pataky J (1989) Smut gall development in adult corn plants inoculated with Ustilago maydis. Plant Dis 73:921–925
Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310:1031–1034
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006
Veldboom LR, Lee M, Woodman WL (1994) Molecular marker-facilitated studies in an elite maize population: I. Linkage analysis and determination of QTL for morphological traits. Theor Appl Genet 88:7–16
Wang C, Yang Q, Wang W, Li Y, Guo Y, Zhang D, Ma X, Song W, Zhao J, Xu M (2017) A transposon-directed epigenetic change in ZmCCT underlies quantitative resistance to Gibberella stalk rot in maize. New Phytol. doi:10.1111/nph.14688
Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol 164:735–747
Wilken GC (1990) Good farmers: Traditional agricultural resource management in Mexico and Central America. Univ of California Press, California
Wu W, Zheng X-M, Lu G, Zhong Z, Gao H, Chen L, Wu C, Wang H-J, Wang Q, Zhou K (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Nat Acad Sci 110:2775–2780
Xu G, Wang X, Huang C, Xu D, Li D, Tian J, Chen Q, Wang C, Liang Y, Wu Y, Yang X, Tian F (2017) Complex genetic architecture underlies maize tassel domestication. New Phytol 214:852–864
Yan J, Yang X, Shah T, Sánchez-Villeda H, Li J, Warburton M, Zhou Y, Crouch JH, Xu Y (2010) High-throughput SNP genotyping with the GoldenGate assay in maize. Mol Breed 25:441–451
Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, Liu T, Liu H, Zhang C, Zhang Z (2013) Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res 23:969–971
Yang Q, Yin G, Guo Y, Zhang D, Chen S, Xu M (2010) A major QTL for resistance to Gibberella stalk rot in maize. Theor Appl Genet 121:673–687
Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Li J, Yan J (2011) Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed 28:511–526
Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T (2013) CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Nat Acad Sci 110:16969–16974
Yang S, Murphy RL, Morishige DT, Klein PE, Rooney WL, Mullet JE (2014a) Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS One 9(8):e105352
Yang S, Weers BD, Morishige DT, Mullet JE (2014b) CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol 14:148
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12(12):2473–2483
Yin C, Li H, Li S, Xu L, Zhao Z, Wang J (2015) Genetic dissection on rice grain shape by the two-dimensional image analysis in one japonica × indica population consisting of recombinant inbred lines. Theor Appl Genet 128:1969–1986
Zhang L, Li Q, Dong H, He Q, Liang L, Tan C, Han Z, Yao W, Li G, Zhao H (2015) Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci Rep 5:7663
Zhao X, Tan G, Xing Y, Wei L, Chao Q, Zuo W, Lübberstedt T, Xu M (2012) Marker-assisted introgression of qHSR1 to improve maize resistance to head smut. Mol Breed 30:1077–1088
Zhong Q, Zhou P, Yao Q, Mao K (2009) A novel segmentation algorithm for clustered slender-particles. Comput Electron Agric 69:118–127
Acknowledgements
We thank Chunfang Ji, Lijun Ren, and Guoqing Tan for their assistance in field management. This work was financially supported by the Ministry of Agriculture of China (Grant number 2016ZX08009-003-001) and the key project of Beijing Municipal Science & Technology Commission (Grant number D16110500060000).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dr. Natalia de Leon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Tong, L., Deng, L. et al. Evaluation of ZmCCT haplotypes for genetic improvement of maize hybrids. Theor Appl Genet 130, 2587–2600 (2017). https://doi.org/10.1007/s00122-017-2978-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2978-1