Abstract
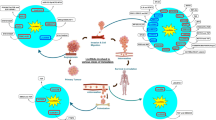
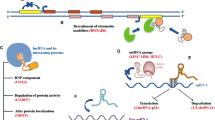
Long non-coding RNA (lncRNA) genes have recently been discovered as key regulators of developmental, physiological, and pathological processes in humans. Recent studies indicate that lncRNAs regulate every step of gene expression, and their aberrant expression can be found in the majority of cancer types. Particularly, lncRNAs were found to function in tumor development and metastasis, which is the major cause of cancer-related death. Thus, exploring key roles of lncRNAs in metastasis is predicted to enhance our knowledge of metastasis, and uncover novel therapeutic targets and biomarkers of this process. In this review, we discuss the molecular mechanisms of lncRNAs in gene expression regulation and their function in metastasis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science, 309, 1559–1563.
Banfai, B., Jia, H., Khatun, J., Wood, E., Risk, B., Gundling Jr., W. E., et al. (2012). Long noncoding RNAs are rarely translated in two human cell lines. Genome Research, 22, 1646–1657.
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature, 543, 199–204.
Harrow, J., Frankish, A., Gonzales, J. M., Tapanari, E., Diekhans, M., Kokocinski, F., et al. (2012). GENCODE: the reference human genome annotation for the ENCODE project. Genome Research, 22, 1760–1774.
Plath, K., Fang, J., Mlynarczyk-Evans, S. K., Cao, R., Worringer, K. A., Wang, H., et al. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science, 300, 131–135.
Pasmant, E., Laurendeau, I., Heron, D., Vidaud, M., Vidaud, D., & Bieche, I. (2007). Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Research, 67, 3963–3969.
Yap, K. L., Li, S., Munoz-Cabello, A. M., Raguz, Z., & Zeng, L. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular Cell, 38, 662–674.
Hu, X., Feng, Y., Zhang, D., Zhao, S. D., Hu, Z., Greshock, J., et al. (2014). A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell, 26, 344–357.
Gumireddy, K., Li, A., Yan, J., Setoyama, T., Johannes, G. J., Orom, U. A., et al. (2013). Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. The EMBO Journal, 32, 2672–2684.
Yoon, J., Abdelmohsen, K., Srikantan, S., Yang, X., Martindale, J. L., De, S., et al. (2012). LincRNA-p21 suppresses target mRNA translation. Molecular Cell, 47, 648–655.
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., & Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465, 1033–1038.
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., & Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science Signaling, 3, ra8.
Fidler, I. J. (2003). The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nature Reviews. Cancer, 3, 453–458.
Yan, J., & Huang, Q. (2012). Genomic screens for metastasis genes. Cancer Metastasis Reviews, 31, 419–428.
Yan, J., Yang, Q., & Huang, Q. (2012). Metastasis suppressor genes. Histology and Histopathology, 28, 285–292.
Chrisholm, K. M., Wan, Y., Li, R., Montgomery, K. D., Chang, H. Y., & West, R. B. (2012). Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One, 7, e47998.
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Research, 71, 6320–6326.
Loewen, G., Jayawickramarajah, J., Zhuo, Y., & Shan, B. (2014). Function of lncRNA HOTAIR in lung cancer. Journal of Hematology & Oncology, 7, 90.
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329, 689–693.
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076.
Ayesh, S., Matouk, I., Schneider, T., Ohana, P., Laster, M., Al-Sharef, W., et al. (2002). Possible physiological role of H19 RNA. Molecular Carcinogenesis, 35, 63–74.
Raveh, E., Matouk, U., Gilon, M., & Hochberg, A. (2015). The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Molecular Cancer, 14, 184.
Liang, W. C., Fu, W. M., Wong, C. W., Wang, Y., Wang, W. M., Hu, G. X., et al. (2015). The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget, 6, 22513–22525.
Luo, M., Li, Z., Wang, W., Zeng, Y., & Liu, Z. (2013). Long noncoding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Letters, 333, 213–221.
Ling, H., Spizzo, R., Atlasi, Y., Nicoloso, M., Shimizu, M., Redis, R. S., et al. (2013). CCAT2, a novel noncoding RNA mapping to 8q24 underlies metastatic progression and chromosomal instability in colon cancer. Genome Research, 23, 1446–1461.
Redis, R. S., Sieuwerts, A. M., Look, M. P., Tudoran, O., Ivan, C., Spizzo, R., et al. (2013). CCAT2, a novel long noncoding RNA in breast cancer: expression study and clinical correlations. Oncotarget, 4, 1748–1762.
Yang, F., Zhang, L., Huo, X. S., Yuan, J. H., Xu, D., Yuan, S. X., et al. (2011). Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology, 54, 1679–1689.
Yang, F., Huo, X. S., Yuan, S. X., Zhang, L., Zhou, W. P., Wang, F., et al. (2013). Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular Cell, 49, 1083–1097.
Hu, Q., Lu, Y. Y., Noh, H., Hong, S., Dong, Z., Ding, H. F., et al. (2013). Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene, 32, 3933–3943.
Liu, B., Sun, L., Liu, Q., Gong, C., Yao, Y., Lv, X., et al. (2015). A cytoplasmic NF-ĸB interacting long noncoding RNA blocks IĸB phosphorylation and suppresses breast cancer metastasis. Cancer Cell, 27, 370–381.
Huang, J. F., Guo, Y. J., Zhao, C. X., Yuan, S. X., Wang, Y., Tang, G. N., et al. (2013). Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression of HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology, 57, 1882–1892.
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031–8041.
Ying, L., Chen, Q., Wang, Y., Zhou, Z., Huang, Y., & Qiu, F. (2012). Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Molecular Biosystems, 8, 2289–2294.
Li, Y., Yang, Z., Wan, X., Zhou, J., Zhang, Y., Ma, H., et al. (2016). Clinical prognostic value of metastasis-associated adenocarcinoma transcript 1 in various human cancers: an updated meta-analysis. The International Journal of Biological Markers, 31, e173–e182.
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell, 39, 925–938.
Wilusz, J. E. (2016). Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochimica et Biophysica Acta, 1859, 128–138.
Gutschner, T., Hämmerle, M., Eissmann, M., Hsu, J., Kim, Y., Hung, G., et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Research, 73, 1180–1189.
Khalil, A. M., Guttman, M., Harte, M., Garber, M., Raj, A., Rivea-Morales, D., et al. (2009). Decreased expression of large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America, 106, 11667–11672.
Xu, T. P., Huang, M. D., Xia, R., Liu, X. X., Sun, M., Yin, L., et al. (2014). Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. Journal of Hematology & Oncology, 7, 63.
Hou, P., Zhao, Y., Li, Z., Ma, M., Gao, Y., Zhao, L., et al. (2014). LincRNA ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death & Disease, 5, e1287.
Eades, G., Wolfson, B., Zhang, Y., Li, Q., Yao, Y., & Zhou, Q. (2015). LincRNA ROR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Molecular Cancer Research, 13, 330–338.
Sun, T. T., He, J., Liang, Q., Ren, L. L., Yan, T. T., Yu, T. C., et al. (2016). LncRNA GClnc1 promotes gastric carcinogenesis and may act as a molecular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discovery, 6, 784–801.
Chang, B., Yang, H., Jiao, Y., Wang, K., Liu, Z., Wu, P., et al. (2016). SOD2 deregulation enhances migration, invasion and has poor prognosis in salivary adenoid cystic carcinoma. Scientific Reports, 6, 25918.
Yuan, J. H., Yang, F., Wang, F., Ma, J. Z., Guo, Y. J., Tao, Q. F., et al. (2014). A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell, 25, 666–681.
Prensner, J. R., Zhao, S., Erho, N., Schipper, M., Iyer, M. K., Dhanasekaran, S. M., et al. (2014). RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SCHLAP1. The Lancet Oncology, 15, 1469–1480.
Mehra, R., Udager, A. M., Ahearn, T. U., Cao, X., Feng, F. Y., Loda, M., et al. (2016). Overexpression of the long non-coding RNA SCHLAP1 independently predicts lethal prostate cancer. European Urology, 70, 549–552.
Prensner, J. R., Iyer, M. K., Sahu, A., Asangani, I. A., Cao, Q., Patel, L., et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature Genetics, 45, 1392–1398.
Meijer, D., van Agthoven, T., Bosma, P. T., Nooter, K., & Dorssers, L. C. (2006). Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Molecular Cancer Research, 4, 379–386.
Godinho, M. F., Sieuwerts, A. M., Look, M. P., Meijer, D., Foekens, J. A., Dorssers, L. C., et al. (2010). Relevance of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. British Journal of Cancer, 103, 1284–1291.
Xing, Z., Lin, A., Li, C., Liang, K., Wang, S., Liu, Y., et al. (2014). LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell, 159, 1110–1125.
Washington, K., Ammosova, T., Beullens, M., Jerebtsova, M., Kumar, A., Bollen, M., et al. (2002). Protein phosphatase-1 dephosphorylates the C-terminal domain of RNA polymerase II. The Journal of Biological Chemistry, 277, 40442–40448.
Sun, N. X., Ye, C., Zhao, Q., Zhang, Q., Xu, C., Wang, S. B., et al. (2014). Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One, 9, e100340.
Yan, Y., Shen, Z., Gao, Z., Cao, J., Yang, Y., Wang, B., et al. (2015). Long noncoding ribonucleic acid specific for distant metastasis of gastric cancer is associated with TRIM16 expression and facilitates tumor cell invasion in vitro. Journal of Gastroenterology and Hepatology, 30, 1367–1375.
Fang, Z., Wu, L., Wang, L., Yang, Y., Meng, Y., & Yang, H. (2014). Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, 117, 89–95.
Li, J. Y., Ma, X., & Zhang, C. B. (2014). Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. International Journal of Clinical and Experimental Pathology, 7, 7938–7944.
Han, Y., Yang, Y. N., Yuan, H. H., Zhang, T. T., Sui, H., Wei, X. L., et al. (2014). UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology, 46, 396–401.
Xue, M., Li, X., Li, Z., & Chen, W. (2014). Urothelial carcinoma associated 1 is a hypoxia inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biology, 35, 6901–6912.
Wang, Z., He, C., Hu, L., Shi, H., Li, J., Gu, Q., et al. (2017). Long noncoding RNA UCA1 promotes tumor metastasis by inducing GRK2 degradation in gastric cancer. Cancer Letters, 408, 10–21.
Zhou, Y., Wang, X., Zhang, J., He, A., Wang, Y., Han, K., et al. (2017). Artesunate suppresses the viability and mobility of prostate cancer cells through UCA1, the sponge of miR-184. Oncotarget, 8, 18260–18270.
Cai, Q., Jin, L., Wang, S., Zhou, D., Wang, J., Tang, Z., et al. (2017). Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget, 8, 47957–47968.
Li, D., Li, H., Yang, Y., & Kang, L. (2017). Long noncoding RNA urothelial carcinoma associated 1 promotes the proliferation and metastasis of human lung tumor cells by regulating microRNA-144. Oncology Research. https://doi.org/10.3727/096504017X15009792179602.
Shalem, O., Sanjana, N. E., & Zhang, F. (2015). High-throughput functional genomics using CRISPR-Cas9. Nature Reviews. Genetics, 16, 299–311.
Korkmaz, G., Lopes, R., Ugalde, A. P., Nevedomskaya, E., Han, R., Myacheva, K., et al. (2016). Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nature Biotechnology, 34, 192–198.
Funding
This study was supported by NIH/NCI R01CA148759, NIH/NCI R21 CA198558-01A1, NIH/NCI U01 CA200495-01, Pardee Foundation, PA Breast and Cervical Cancer Research Initiative, and National Natural Science Funds (81372328, 81572834).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Huang, Q., Yan, J. & Agami, R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev 37, 75–81 (2018). https://doi.org/10.1007/s10555-017-9713-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-017-9713-x