Abstract
When embryos experience different environments than their parents, plasticity can enable the development of alternate phenotypes that confer higher fitness in the new conditions. Temperature-induced plasticity could be especially critical for species that inhabit areas with considerable thermal variation. We studied transcriptional variation in embryos of mangrove rivulus (Kryptolebias marmoratus)—a self-fertilizing hermaphroditic, eurythermal fish that resides in notoriously spatiotemporally variable mangrove forests—exposed to different thermal regimes during development. To study transcriptional plasticity, we first improved the genome assembly to chromosome length scaffolds (N50 of 28.17 Megabases). Whole transcriptome sequencing revealed that both temperature and developmental timing modulated embryonic gene expression. We found few differences in gene expression between embryos incubated in cold and warm conditions and assessed before the temperature-sensitive period of development, indicating high resistance to stochastic changes in gene expression early in development. Replicate embryos exposed to cold temperatures and sampled after the temperature-sensitive period showed less variation in gene expression than those sampled before, suggesting canalization of the plastic response. DNA replication/repair, organelle, and gas transport pathways were upregulated while nervous system development, cell signaling, and cell adhesion were downregulated in cold-exposed compared to warm-exposed embryos sampled after the temperature-sensitive period. These plastic shifts in gene expression could have major implications for reorganizing the phenotype (e.g., apoptosis, mitosis) in response to environmental changes occurring within a generation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Avoid common mistakes on your manuscript.
Introduction
An individual’s phenotype arises from the interplay between its genotype, the environment, and their interactions, resulting in traits that range from stable to variable. Invariant traits are insensitive to the environment and show little variance among genetically identical individuals, while variable traits differ stochastically between individuals with the same genotype occupying the same environment (Abley et al. 2016). Plastic traits, however, are highly environmentally dependent, with phenotypic variance being significantly lower among individuals occupying (or raised in) the same environment than in different environments. The expression of plastic traits and their sensitivity to environmental inputs also vary, often predictably, among genotypes (genotype-by-environment interactions). Plasticity has been observed across taxa in response to both biotic and abiotic stimuli such as changes in predator abundance, light, precipitation, and temperature (Hansson 2004; Härer et al. 2017; Mallard et al. 2020; Sentis et al. 2017). Plasticity often is adaptive because it enables relatively rapid responses to environmental change. However, plasticity can also be maladaptive, particularly in cases where environments experienced during development and adulthood are incongruent (Cenzer 2017; Langerhans and DeWitt 2002). In this sense, plasticity can have major evolutionary implications by altering the distribution of phenotypes (e.g., shifting trait values closer to or further away from some theoretical “optimum” with highest relative fitness), thereby protecting alleles from or exposing alleles to natural selection (Fox et al. 2019; Huey et al. 2003). Understanding the mechanisms underlying plastic responses to the environment, which can produce considerable phenotypic diversity within a species, is essential for evaluating species-specific evolutionary responses to environmental change.
Given that climate change will continue to alter thermal regimes (i.e., temporal patterns of temperature change on any scale), exploring phenotypic responses to changes in temperature is particularly timely. Multiple animal taxa have temperature-induced plasticity that contributes to observed phenotypic variation (Gleason et al. 2018; Orizaola and Laurila 2008; Sawall et al. 2015). The ability of an individual to mount a plastic response to temperature often depends on the life stage during which the individual is exposed (Fraimout et al. 2018; Noble et al. 2018; While et al. 2018), often called thermocritical or thermolabile periods. Such plastic responses to temperature can be accompanied by pronounced changes in gene expression that presumably dictate subsequent shifts in the development of the phenotype (Riddell et al. 2019; Zhu et al. 2016). The interaction between thermal regime and developmental stage could thus drive plastic changes in gene expression that have important ecological and evolutionary consequences.
Species within intertidal habitats (e.g., rocky intertidal, mangrove forests) often experience significant variations in water temperature both temporally (Mislan et al. 2009; Morris and Taylor 1983; Wolfe et al. 2020) and spatially (Helmuth et al. 2016; Serôdio and Catarino 1999). Spatial variation in temperature accompanies changes in microhabitat within the intertidal, which can be driven by both abiotic (Denny et al. 2006; Helmuth and Hofmann 2001; Miller et al. 2009) and biotic factors (Guo et al. 2017; Taylor 1992). Therefore, species within intertidal habitats are ideal models in which to explore time-sensitive, temperature-dependent phenotypic plasticity. In these environments, temperature fluctuations across both time and space are ecologically relevant (e.g., can dictate community structure) and likely to become more extreme in the coming decades (Garcia et al. 2014; He and Silliman 2019; IPPC 2014). Moreover, tropical species are currently experiencing more cold extremes, and the incidence is likely to accelerate with time (Williams et al. 2015; Leriorato et al. 2021).
The mangrove rivulus fish (Kryptolebias marmoratus; henceforth “rivulus”) is a small, cryptic, eurythermal fish inhabiting New World mangrove forests of Central America, the Bahamas, peninsular Florida (USA), and the Florida Keys (USA) (Tatarenkov et al. 2017; Taylor 2012). Rivulus populations consist mostly of hermaphrodites and very few males (typically < 5% but up to 25% in Twin Cayes, Belize; Davis et al. 1990; Turner et al. 1992). They reproduce primarily through self-fertilization (“selfing”), although rare outcrossing events occur (Harrington 1961; Mackiewicz et al. 2006; Tatarenkov et al. 2015; Tatarenkov et al. 2017). After repeated generations of selfing, rivulus can produce isogenic lineages with offspring nearly genetically identical to both the parent and siblings (Harrington 1968), with some heterozygosity persisting (Lins et al. 2018). Rivulus also are quite sensitive to temperature during embryonic development. When eggs are exposed to 18–20 °C at the beginning of the thermolabile period (embryonic developmental stage 31; Mourabit et al. 2011), a significant proportion of individuals develop into males, with the exact proportion being genotype-dependent (Ellison et al. 2015; Harrington 1968; Turner et al. 2006). Temperature and its interaction with genotype also drive changes in growth rate, with genotypes varying in the extent to which growth rates decrease in low temperatures (Lin and Dunson 1999). Rivulus are often found in warm water (> 20°; Turner et al. 2006) but survive in the wild at temperatures between 7 and 38 °C (Taylor et al. 1995), making temperature a biologically relevant stressor that drives plastic responses. Rivulus is thus an appropriate model in which to investigate time-sensitive, temperature-induced plasticity in gene expression because of the following: (i) they exhibit phenotypically plastic responses to temperature; (ii) these plastic responses are initiated during specific developmental periods; and (iii) isogenic lineages can be used to control for genotype and disentangle environmental effects.
Using previously developed -omics resources (Kelley et al. 2012; 2016), we investigated how embryonic exposure to divergent thermal regimes alters whole transcriptome patterns of gene expression. We also scaffolded rivulus’ genome, providing a chromosome length reference for studies seeking to investigate the molecular underpinnings of temperature-induced plasticity. We aimed to determine which molecular pathways change in rivulus embryos incubated in cold (20 °C) or warm (25 °C) water, as well as before and after the thermolabile period (stage 31) in each temperature. We hypothesized that variation in gene expression would exist between temperature treatments (20 °C, 25 °C) both before and after the thermolabile period. We predicted that the most dramatic temporal changes in gene expression would correspond to cold temperature treatments, given that both sex and growth rates are significantly altered by low temperature exposure during embryonic development (Ellison et al. 2015; Harrington 1968; Lin and Dunson 1999; Turner et al. 2006).
Materials and methods
Experimental design
We used one isogenic lineage of Kryptolebias marmoratus collected by D. Scott Taylor from Reckley Hill Lake, San Salvador, Bahamas in 1997 and maintained in the Earley lab since 2008. We focused on this specific lineage (hereafter, RHL) because it was used to generate the reference genome (Kelley et al. 2016; Lins et al. 2018), on which this study expands. Adults were housed individually in ventilated 1.2 L Rubbermaid® containers (TakeAlongs® deep square) filled with 600 ml of 25 parts per thousand (ppt) salt water (aged tap water and Instant Ocean® salt) and maintained on a 12-h light:12-h dark photoperiod. Ambient (air) temperature was 26.2 ± 0.002 °C (mean ± SEM for temperature recordings every 20 min for the study duration); water temperature is typically 1–2 °C cooler than the air temperature. Fish were fed daily with 2 ml brine shrimp (Artemia) nauplii suspended in 25 ppt water (~ 500 nauplii/ml). Poly-Fil® was placed in the corner of each container at the air–water interface as egg-laying substrate and was checked twice weekly.
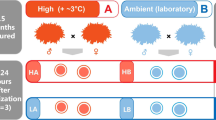
Self-fertilized eggs were collected from each container and photographed with a Canon® G9 camera attached to a Zeiss Stemi 2000-C stereomicroscope to determine the initial developmental stage (Mourabit et al. 2011); fertilized eggs can be retained by the mother for various amounts of time before being laid. Eggs collected prior to developmental stage 20 (Mourabit et al. 2011) were placed individually in 7 ml, 13 × 100 mm polystyrene test tubes filled halfway with 25 ppt saltwater. Tubes were labeled with unique identification numbers and randomly assigned to one of two time points—whether the embryo would be removed before or after the thermolabile period of development (stage 31 [Harrington 1968; Mourabit et al. 2011]), which we refer to as Pre or Post critical period, respectively. Embryos also were assigned to water temperature treatments (20 °C or 25 °C; Cold and Warm, respectively), with the warm (25 °C) condition effectively serving as the control because this is the water temperature at which the fish are normally held. Embryos assigned to Pre were removed at developmental stages 29 or 30, and those assigned to Post were removed at developmental stage 32 (see Mourabit et al. 2011). Time point assignments (Pre and Post) were crossed in a full-factorial design with temperature treatment to generate four comparison groups: Pre-Cold (PrC), Post-Cold (PoC), Pre-Warm (PrW), Post-Warm (PoW). Tubes containing embryos were then placed haphazardly into weighted Bel-Art™ No-Wire grip racks within the center of a water bath set to the assigned temperature-controlled treatment. A formula was developed based on the timing of development described in Mourabit et al. (2011) to provide an estimate of the date each embryo would reach the thermolabile period (stage 31; Supporting Information (SI): Developmental Formula) using the initial development stage and the number of hours developing. The timing of visual inspections of the eggs was also based on temperature treatment (e.g., embryos in cold water develop slower than embryos in warm water). Thus, embryos were exposed to their respective temperature treatment from the time of egg collection (with documented stage of development) to either Pre or Post, depending on time point assignment.
Treatment set-up and treatment details
Treatment tanks of temperature-controlled water were kept circulating by ViaAqua® 350 and Marineland® Maxi-Jet 600 submersible pumps. The Cold tank was kept at 20 °C with an Arctica® Titanium Chiller (1/10 HP). The Warm tank had water pumped in using a Marineland® Maxi-Jet 600 submersible pump from a separate bucket chilled with a NOVA TEC IceProbe® Water Chiller through a Hydor™ ETH In-Line Heater to maintain the temperature at 25 °C. Temperature was monitored with Alpha Mach iBCod 22L temperature data loggers that were placed in a central location within each tank (SI Table 1). Water was added to each tank approximately twice per week to always maintain water levels near the top of the tubes, ensuring eggs were always in treatment. Water temperature in the tanks was measured before and after the replacement of evaporated water to ensure that the temperature did not change more than one degree, which could be regulated by the heaters and chillers quickly. In each tank, an Erlenmeyer flask with 25 ppt salt water was kept at the appropriate temperature to ensure that eggs were immediately exposed to that temperature when first loaded and for each weekly water change.
Final staging and processing
Once an embryo reached the appropriate developmental stage, estimated using the aforementioned formula (SI), it was gently removed from treatment with a transfer pipette, placed on a petri dish, and photographed with the Canon® G9 camera attached to a Zeiss Stemi 2000-C stereomicroscope. Developmental stage was determined by comparing the photograph to Mourabit et al. (2011). Embryos were transferred directly to a 0.5-ml microcentrifuge tube filled with RNAlater. Labeled microcentrifuge tubes were kept at 4 °C for 24 h then stored at –80 °C.
Genome improvement
Library preparation and sequencing
To improve the existing reference genome, a Chicago library was prepared as described by Putnam et al. (2016). Briefly, ~ 500 ng of High Molecular Weight gDNA (mean fragment length = 75 kilobases (kb)) was reconstituted into chromatin in vitro and fixed with formaldehyde. Fixed chromatin was digested with DpnII, the 5’ overhangs filled in with biotinylated nucleotides, and free blunt ends ligated. After ligation, crosslinks were reversed, and DNA purified from protein was treated to remove biotin external to ligated fragments. DNA was then sheared to ~ 350 basepairs (bp) mean fragment size, and sequencing libraries were generated using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. Libraries were sequenced on an Illumina HiSeq X to produce 242 million 2 × 150 bp paired-end reads, which provided 176.60 × physical coverage of the genome (1–100 kb). Two Dovetail HiC libraries were prepared as described for the Chicago library (Lieberman-Aiden et al. 2009). The number and length of read pairs produced for each library were as follows: 40 million, 2 × 150 bp for library 1; 178 million, 2 × 150 bp for library 2. Together, these Dovetail HiC library reads provided 86.97 × physical coverage of the genome (10–10,000 kb).
Scaffolding the assembly with HiRise
The draft de novo assembly (Rhee et al. 2017), short reads, Chicago library reads, and Dovetail HiC library reads were used as input data for HiRise, a software pipeline designed specifically for using proximity ligation data to scaffold genome assemblies (Putnam et al. 2016). An iterative analysis was conducted. First, shotgun and Chicago library sequences were aligned to the draft input assembly using a modified SNAP read mapper (http://snap.cs.berkeley.edu). The distance between Chicago read pairs mapped within draft scaffolds were analyzed by HiRise to produce a likelihood model for genomic distance between read pairs. The model was used to identify and break putative misjoins, score prospective joins, and make joins. After aligning and scaffolding Chicago data, Dovetail HiC library sequences were aligned and scaffolded in the same way. After scaffolding, shotgun sequences were used to close gaps between contigs.
Completeness of the genome was determined using the program Benchmarking Universal Single-Copy Orthologs (BUSCO) and the general eukaryota_odb10 and specific cyprinodontiformes_odb10 BUSCO databases (Simão et al. 2015). To visualize genome contiguity, genetic markers for the 24 linkage groups identified by Kanamori and colleagues (2016) were mapped to our newly scaffolded genome using BWA (Li and Durbin 2009). The resulting SAM file with a randomly chosen single marker per unique map position was used in R (version 4.0.3) to create the datafile for Fig. 1.
Visualization of the relationship between the improved genome assembly from this study and previously described linkage groups (Kanamori et al. 2016), demonstrating chromosome length scaffolding. The figure includes the N50 (28.17 Mb) and BUSCO-related counts and percentages included for both eukaryotes (98.9%) and Cyprinodontiformes (91.9%)
RNA isolation and RNA-sequencing library preparation
Twelve embryos, three per treatment, were shipped to Washington State University and stored at − 80 °C. RNA was isolated using the Macherey–Nagel NucleoSpin RNA XS kit according to the manufacturer’s instructions. RNA quantity and quality were determined using a Qubit fluorometer and an Advanced Analytical Technologies (AATI) Fragment Analyzer, respectively. High-quality samples were normalized to 500 ng total RNA for input into library preparation. Samples were enriched for mRNA using New England Biolabs NEBNext Poly(A) mRNA Magnetic Isolation Module and prepared for sequencing using NEBNext Ultra Directional RNA Library Prep Kit for Illumina. To determine the molarity of each library, samples were run on an AATI Fragment Analyzer. An equal molar pool was created from all libraries and sequenced on one lane of an Illumina HiSeq 2500 System (paired end, 125 bp reads) at the New York Genome Center. All sequence data were deposited in the Sequence Read Archive on GenBank (BioProject ID: PRJNA401942).
Read mapping and gene expression matrix
Raw reads generated from the libraries above were adapter trimmed and then quality trimmed using Trim Galore! (v0.4.1) (Krueger 2015). Trimmed reads were subsampled so that each set of paired reads would not exceed a total of 20 million reads. Trimmed reads were mapped to the newly scaffolded Kryptolebias marmoratus genome (GenBank Accession: GCF_ 001649575.2) using Hisat2 (version 2.1.0), which was guided by reference annotation (Kim et al. 2015). Resulting SAM files had mate pair information fixed using Picard Tools FixMateInformation (version 2.21.4) and converted to BAM files using SAMtools (version 1.2) view (Li et al. 2009). StringTie (version 1.3.4 d) (Pertea et al. 2015) generated gene counts for each embryo guided by the reference annotations. A Python script included in the StringTie download, prepDE.py, merged gene counts for each embryo into a gene count matrix.
Gene expression patterns
Henceforth, all treatments are referred to by their acronym: PrC (Pre-Cold), PoC (Post-Cold), PrW (Pre-Warm), PoW (Post-Warm). The gene count matrix was loaded into R (v 4.0.3) (R Core Team 2020) and genes with fewer than 0.5 counts per million in three individuals were filtered out. To visualize similarity or dissimilarity in gene expression patterns between samples, we plotted the top 500, 1000, and 10,000 genes based on log2-fold change on a multidimensional scaling (MDS) plot using pairwise comparisons and the package limma (Ritchie et al. 2015; SI Figs. 2–4).
Differential expression was quantified using the Bioconductor package edgeR (version 3.32.1), implementing a generalized linear model (GLM) (Robinson et al. 2010). To test for statistical significance, we used the GLM quasi-likelihood F-test (glmQLFTest in edgeR) with a Benjamini–Hochberg correction. Significance was set at a false discovery rate (FDR) < 0.05. To disentangle how developmental stage, temperature, and their interaction influenced gene expression, we performed a series of pairwise comparisons. To determine gene expression changes that could be related to temperature, we compared PrC to PrW embryos and PoC to PoW embryos. To determine changes in gene expression across development at a specific temperature, we compared PrC to PoC embryos and PrW to PoW embryos.
Gene ontology and Kyoto encyclopedia of genes and genome enrichment
Gene ontology (GO) analyses were performed for all comparisons with significant differentially expressed genes (PrC vs. PrW, PrC vs. PoC, PoC vs. PoW but not PrW vs. PoW) using the gprofiler2 package (version 0.2.0) (Kolberg et al. 2020). GO terms were found for three ontologies: biological process, cellular component, and molecular function. To determine which GO terms were significant, a one-sided Fisher’s exact test with FDR < 0.05 was performed separately for significantly upregulated and downregulated genes in each comparison using all annotated genes as the background (e.g., PrC vs. PrW). Similarly, Kyoto encyclopedia of genes and genome (KEGG) enrichment analyses at the level of KEGG ontology and pathway were performed for the same comparisons using clusterProfiler (Xu et al. 2024) using a Fisher’s exact test with FDR < 0.05 with a minimum gene set size of five. While gprofiler2 can also test for KEGG enrichment, the package only uses precompiled data and KEGG was not available for rivulus through the package. ClusterProfiler allows for user-defined gene-to-term mapping files. Therefore, clusterProfiler was used with all genes found in our expression matrix as the background set with KEGG terms and ontology extracted from the annotation file.
Results
Genome scaffolding and annotation
Chicago library preparation and sequencing generated 242 million, 150 bp paired-end reads, providing 263.57 × coverage of the genome. After Chicago sequencing, the reference genome (GCF_001649575.1) was improved from a scaffold N50 of 2.23 Megabases (Mb) to 5.02 Mb. After Dovetail HiRise assembly, the genome was further improved to a scaffold N50 of 28.17 Mb. The total number of scaffolds was reduced from 3073 to 300, including the mitochondrial genome. Of the 299 nuclear scaffolds, 27 contained at least one genetic marker from the previously published genetic map for K. marmoratus (Kanamori et al. 2016). Of the 27 scaffolds with at least one marker, more than 99% of the genetic markers were contained within 24 of those scaffolds, indicating that the new genome is scaffolded to chromosome length (Fig. 1), with the additional three scaffolds possibly being difficult-to-assemble regions of the chromosomes. Rivulus has a haploid karyotype of n = 24 (Scheel 1972). The newly scaffolded genome had 252 of 255 eukaryotic complete BUSCOs (98.9%). A total of 15,213 Cyprinodontiformes BUSCOs were searched, and there were 14,024 (92.2%) complete BUSCOs, including 13,976 (91.9%) single copy and 48 (0.3%) complete and duplicated, as well as 126 (0.8%) fragmented BUSCOs in the newly scaffolded genome (Fig. 1).
Embryo RNA
Total trimmed reads ranged from 7,176,142 to 60,088,608 and 7,176,142 to 40,000,000 after subsampling. Mapping percentages ranged from 42.5 to 91.6% (SI Table 2). Two samples from the PrW treatment had low mapping percentages (SI Table 2) that could potentially impact our results. However, the last PrW sample had the highest percent of reads mapped (91.6%; SI Table 2), and our filtering regime removes genes expressed at low levels (less than 0.5 counts per million in three samples). Total counts for differential expression analysis, prior to filtering, ranged from 2,452,255 (RPRW_112) to 35,119,175 (RPRW_39). After filtering for genes expressed at a low level (less than 0.5 counts per million in at least three individuals), 21,447 of the 25,186 genes remained.
Gene expression patterns
MDS plots for the 500, 1000, and 10,000 genes with the greatest log fold change showed considerable separation between treatments (SI Figs. 1–3), the most prominent being between PoC embryos and embryos from the remaining treatments (PrC, PrW, PoW). Generally, samples from each treatment were distinct from other treatments, regardless of the number of genes selected (500, 1000, 10,000), indicating similar gene expression profiles among replicates. PoC and PrW samples, however, were most highly clustered, suggesting relatively low within-treatment variation compared to PrC and PoW (SI Figs. 1–3).
The vast majority of significantly differentially expressed genes occurred between embryos sampled before and after the thermolabile period in cold temperatures (PrC vs. PoC; 1,313 with ~ 75% being downregulated in PrC relative to PoC) and between embryos sampled after the thermolabile period in the two different temperatures (PoC vs. PoW; 1,570 with an approximately equivalent number of up- and downregulated genes) (Table 1). There were zero significantly differentially expressed genes between embryos sampled before and after the thermolabile period in warm temperatures (PrW vs. PoW), and only 43 between embryos sampled before the thermolabile period in the two different temperatures (PrC vs. PrW) (Table 1, SI Table 3, SI Fig. 4).
Gene ontology and KEGG enrichment
Comparing embryos raised in the cold before and after the thermolabile period (PrC vs. PoC), 286 GO terms, 126 KEGG terms, and 188 KEGG pathways were enriched, with 157, 28, and 63 upregulated along with 129, 98, and 125 downregulated in PrC relative to PoC, respectively (Table 1 (see additional details in SI Table 4, SI Table 5, and SI Table 6)). Upregulated GO terms were associated with neural and sensory development (e.g., differentiation of brain regions [forebrain, thalamus, habenula], retina layer formation, neurogenesis), cell division and differentiation (e.g., G2/M transition of mitotic cycle, microtubule binding, cell fate commitment, transcription factor activity), and macromolecule synthesis, particularly nucleic acids. Upregulated KEGG terms included similar functions such as nervous system development (e.g., HES5, POU-domain transcription factors) and cell division (e.g., STAG1/2). Upregulated KEGG pathways included pathways associated with the cell cycle (e.g., p53 signaling pathway, apoptosis). Downregulated GO terms were diverse and related to intracellular signaling (e.g., calcium ion binding, voltage-gated sodium channels, ion transport), cell–cell connectivity (e.g., desmosomes, adhesion, cell junction organization), vision (e.g., photoreceptor activity, cone photoresponse recovery), energy production (e.g., fatty acid biosynthesis and metabolism, glycolytic process), muscle function (e.g., sarcomere, myofibril, Z-disc), and protein and DNA catabolism (e.g., endopeptidase activity, purine metabolism). Downregulated KEGG terms and pathways aligned well with the observed enriched GO terms, with KEGG terms enriched for visual processes (e.g., GRK1) and muscle functions (e.g., PVALB) alongside KEGG pathways such as phototransduction, cell adhesion, and cardiac muscle contraction.
Comparing embryos raised in cold and warm temperatures before the thermolabile period (PrC vs. PrW), 89 GO terms, 10 KEGG terms, and 31 KEGG pathways were enriched, with 39, 0, and 7 upregulated along with 55, 10, and 21 downregulated GO terms, KEGG terms, and KEGG pathways, respectively, in PrC relative to PrW. Upregulated GO terms fell into two main categories: epigenetic processes (e.g., demethylase activity, H3K27me3 modified histone binding) and gas transport (e.g., heme biosynthesis, oxygen binding, oxygen carrier activity). Upregulated KEGG pathways showed similar trends with the polycomb repressive complex pathway (related to histone modification) and porphyrin metabolism pathway (related to respiration) significantly upregulated. Downregulated GO terms aligned primarily with sensory processes, specifically in the eye (e.g., photoreceptor activity, phototransduction, sensory perception of light), G-protein-related signal transduction (e.g., β/γ-subunit complex binding, adenylate cyclase signaling pathway, small GTPases [Ras/Rho]), and energy utilization (e.g., phosphagen biosynthesis and metabolism, lactate dehydrogenase activity, response to sugars). Downregulated KEGG terms were related to signal transduction (e.g., UNC-119) and photoreceptors (e.g., gamma-crystallin), while downregulated KEGG pathways were related to eye function (e.g., phototransduction), metabolism (e.g., propanoate metabolism, pyruvate metabolism), and intracellular signaling (e.g., ErbB signaling pathway).
Comparing embryos raised in cold and warm temperatures after the thermolabile period (PoC vs. PoW), 141 GO terms, 88 KEGG terms, and 234 KEGG pathways were enriched, with 73, 26, and 133 upregulated, respectively, along with 68, 62, and 101 downregulated in PoC relative to PoW. Upregulated GO terms were predominantly (~ half) associated with DNA replication and repair, and the cell cycle (e.g., telomere maintenance, Ku70:Ku80 complex [DNA repair], recombination, duplex unwinding, cell cycle checkpoint signaling, organelle production). Gas transport (e.g., oxygen binding, hemoglobin complex) also was upregulated in PoC relative to PoW. Upregulated KEGG terms aligned well with the GO term results with terms related to gas transport (e.g., hemoglobin subunit alpha) as well as terms related to the cell cycle (e.g., tubulin alpha, Cdc42). Upregulated KEGG pathways were related to DNA replication and repair (e.g., DNA replication, mismatch repair, nucleotide excision repair) as well as the cell cycle (e.g., p53 signaling pathway, cell cycle). Downregulated GO terms were associated with intracellular and cell–cell signaling (e.g., cation channel activity, calmodulin-dependent protein kinase activity, vesicular or granular secretion, regulation of neurotransmitter levels) and neural function (e.g., neurexin protein binding, pre- and post-synaptic membranes, circadian and locomotor rhythms). Downregulated KEGG terms were related to neural function (e.g., SV2, KChIPs, neurabin, AMIGO proteins) and cell signaling (e.g., PLC-β). Downregulated KEGG pathways included signal pathways (e.g., MAPK signaling pathway, ErbB signaling pathway, ABC transporters).
GO terms, KEGG terms, and KEGG pathways were not evaluated for embryos raised in the warm and sampled before and after the thermolabile period (PrW vs. PoW) because there were no differentially expressed genes between the two treatments.
Discussion
The RNA-sequencing results supported our hypothesis that patterns of gene expression would vary as a function of both temperature (cold vs. warm) and developmental timing (before vs. after thermolabile period). Cold exposure drove significant molecular, cellular, and neural reorganization across development that was lacking in warm-exposed embryos. This temperature-dependent divergence in gene expression was evident after the thermolabile period as well. Cold-exposed embryos invested more in building cells and tissues and transporting oxygen to them, and less in intra- and intercellular communication and central nervous system function than warm-exposed embryos during the final stages of development. We showed a significant number of up- and downregulated transcripts upon cold exposure, which corresponds to findings in another tropical fish (zebrafish; Scott and Johnston 2012) but deviates from the predominant upregulation of genes that occurs with cold exposure in fishes that occupy cooler water (Chen et al. 2008; Gracey et al. 2004). Importantly, variation in gene expression patterns emerged despite using animals from a single isogenic lineage (RHL), which effectively eliminates genotype or genotype-by-environment as sources of molecular variance. The ability to isolate these molecular responses to the environment makes rivulus a useful model to investigate the consequences of environmental change. Future studies, however, should capitalize on the diversity of genotypes that exist within and among populations to determine whether and to what extent our findings are generalizable among lineages. This is particularly important given that rivulus lineages can vary, sometimes considerably, in their critical thermal maxima and in the threshold temperature that elicits emersion from the water (e.g., Currie & Tattersall 2018; Brown et al. 2024). Such variation implies that the scope of plasticity might also differ among lineages; e.g., lower CTmax would narrow the scope, and perhaps have implications for how embryo development changes at different temperatures.
Our results reveal considerable plasticity in gene expression within a lineage, especially during cold exposure, and likely driven by a host of epigenetic processes upregulated before the thermolabile period (in PrC relative to PrW) (see also Ellison et al. 2015). This corresponds to our prediction that cold temperatures would drive drastic changes in gene expression, perhaps to reorganize development in ways that produce dramatically different sexual phenotypes, growth rates, and body size relative to those that develop in warmer conditions (Ellison et al. 2015; Harrington 1968; Lin and Dunson 1999; Turner et al. 2006). By also assembling a chromosome-level genome (Fig. 1), we facilitate future research in this area while opening new potential avenues of research and further developing rivulus as a potential model system.
Temperature regime dependent gene expression patterns
Using a full factorial design, we disentangled the impact of exposure timing and temperature on gene expression patterns across the transcriptome. Because zero genes were differentially expressed between PrW and PoW, warm temperatures are likely a benign environmental scenario. However, differentially expressed genes were observed in all other comparisons (PrC vs. PrW, PoC vs. PoW, and PrC vs. PoC; Table 1). Tight MDS sample clustering of PrW embryos (SI Figs. 2–4) indicates low variability between samples, suggesting that gene expression is tightly regulated in benign conditions before the thermolabile period. Furthermore, PrC vs. PrW treatments had substantially fewer differentially expressed genes (43 genes) relative to the PoC vs. PoW (1570) or PrC vs. PoC (1313) comparisons (Table 1). The transcriptome might thus be more resistant to stochastic or environmentally induced variation in gene expression before the thermolabile period, as previously demonstrated in Drosophila during early development (Liu et al. 2020). PrC samples also were tightly clustered in the MDS plots (SI Figs. 2–4), indicating a predictable developmental response to cold even before the thermolabile period.
Thermally induced plasticity in gene expression during development is well documented (Oliver et al. 2013; Metzger and Schulte 2018); however, our data also revealed the importance of exposure timing. The PrC vs. PoC comparison captured changes across the thermolabile period when embryos were incubated in the cold. Cold temperatures altered gene expression regardless of when exposure took place, but exposure during the thermolabile period drove a pulse of change in the transcriptome (Table 1). Epigenetic processes stood out as being strongly modulated by temperature (e.g., in PrC vs. PrW), with cold exposure upregulating genes encoding proteins that both facilitate (e.g., demethylase) and inhibit (e.g., H3K27me3) downstream gene expression. This could deconstruct and reconstruct the epigenetic landscape well after global methylation has plateaued during embryogenesis, which is already quite late in rivulus relative to other fishes (Fellous et al. 2018; Wang & Bhandari 2020). Thus, cold exposure might open up a window, via epigenetic reprogramming, for wholesale changes in the phenotype (e.g., Fellous et al. 2022); for instance, cold-induced development of males in rivulus (Ellison et al. 2015; Harrington 1968).
Gene ontology and KEGG enrichment
Many of the terms downregulated in PrC relative to PoC also were downregulated in PoC relative to PoW, suggesting that cold temperatures might have the overall effect of slowing development, particularly with respect to cell–cell communication and connectivity, sensory systems, and molecular processes involved in protein and DNA catabolism. While we did not closely monitor the timing of development (and so, cannot capture nonlinear patterns), we knew the stage at which the embryo was collected and the stage when it was removed from the experiment, plus the total hours spent incubating. Embryos incubated in cold took about twice as long to develop as those incubated in warm temperatures (average number of hours per developmental stage; cold: 67.09 ± 13.05, warm: 33.19 ± 5.14). Thus, it is possible that some differentially expressed genes were related to differences between temperature treatments in the absolute time embryos spent incubating.
Interestingly, though, cold-exposed (but not warm-exposed) embryos showed variation across the thermolabile period in expression of genes related to brain and sensory development (higher in PrC than PoC). At the thermolabile period (embryonic developmental stage 31), most organ formation in rivulus is complete (Mourabit et al. 2011). Having more differentially expressed genes before stage 31 (PrC) is thus not entirely unexpected, as those transcripts might ultimately support the final stages of nervous system development. However, warm-exposed embryos showed no such temporal change in gene expression, indicating an interaction between temperature and timing. It is possible that we missed an important transition in transcriptomic activity by sampling warm-exposed embryos too late (e.g., before the thermolabile period but after brain development ceased). Functionally, it could be that cold exposure before the thermolabile period upregulates genes that promote restructuring of the brain and associated signaling processes in ways that have predictable (but perhaps not adaptive) phenotypic outcomes, e.g., behavioral variation. Exposure to low temperatures is associated with decreased short-term memory (Jones et al. 2005) and decreased neuronal growth (Peng et al. 2007) in other species. Rivulus depend on intermittently dry complex microhabitat (Taylor 2012) and often fight with conspecifics (Hsu et al. 2008). Therefore, decreased cognitive and locomotory abilities associated with lower developmental temperatures might disrupt rivulus’ capacity to navigate both its physical and social environments.
Conclusion
Understanding thermally induced plasticity in gene expression is critical for predicting organismal responses to increasingly unpredictable temperature fluctuations (IPCC 2014). Using an isogenic lineage of rivulus, we controlled for genotype and demonstrated that both temperature and developmental stage at exposure drive plastic changes in gene expression. The transcriptome appeared largely resistant to environmental influences before the thermolabile period, and cold temperatures canalized gene expression compared to warmer conditions. Cold-exposed embryos ultimately (after thermolabile period) showed lower gene expression related to neural function and signaling than warm-exposed counterparts, which could have major implications for the behavioral phenotype and individual fitness. Future research should incorporate time and duration of exposure with longitudinal physiology, morphology, and behavioral studies to elucidate how thermal regimes shape individual phenotypes.
Data availability
Raw RNA Sequence data has been deposited under NCBI BioProject PRJNA401942. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LWHD00000000. The version described in this paper is version LWHD02000000. All code is available at the GitHub repository anthonysnead/Plastic-Gene-Rivulus (https://github.com/anthonysnead/Plastic-Gene-Rivulus).
References
Abley K, Locke JCW, Leyser HMO (2016) Developmental mechanisms underlying variable, invariant and plastic phenotypes. Ann Bot 117:733–748. https://doi.org/10.1093/aob/mcw016
Brown S, Rivard GR, Gibson G, Currie S (2024) Warming, stochastic diel fluctuations affect physiological performance and gill plasticity in an amphibious mangrove fish. J Exp Biol 227:jeb246726. https://doi.org/10.1242/jeb.246726
Cenzer ML (2017) Maladaptive plasticity masks the effects of natural selection in the red-shouldered soapberry bug. Am Nat 190:521–533. https://doi.org/10.1086/693456
Chen Z et al (2008) Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci 105(35):12944–12949. https://doi.org/10.1073/pnas.0802432105
Currie S, Tattersall GJ (2018) Social cues can push amphibious fish to their thermal limits. Biol Let 14:20180492. https://doi.org/10.1098/rsbl.2018.0492
Davis W, Taylor DS, Turner B (1990) Field observations of the ecology and habits of mangrove rivulus (Rivulus marmoratus) in Belize and Florida (Teleostei: Cyprinodontiformes: Rivulidae). Ichthyol Explor Freshwaters 1:123–134
Denny MW, Miller LP, Harley CDG (2006) Thermal stress on intertidal limpets: long-term hindcasts and lethal limits. J Exp Biol 209:2420–2431. https://doi.org/10.1242/jeb.02258
Ellison A et al (2015) Epigenetic regulation of sex ratios may explain natural variation in self-fertilization rates. Proc R Soc B: Biol Sci 282:20151900. https://doi.org/10.1098/rspb.2015.1900
Fellous A, Labed-Veydert T, Locrel M, Voisin AS, Earley RL, Silvestre F (2018) DNA methylation in adults and during development of the self-fertilizing mangrove rivulus. Kryptolebias Marmoratus Ecol Evol 8(12):6016–6033. https://doi.org/10.1002/ece3.4141
Fellous A, Wegner KM, John U, Mark FC, Shama LNS (2022) Windows of opportunity: ocean warming shapes temperature-sensitive epigenetic reprogramming and gene expression across gametogenesis and embryogenesis in marine stickleback. Glob Chang Bio 28(1):54–71. https://doi.org/10.1111/gcb.15942
Fox RJ, Donelson JM, Schunter C, Ravasi T, Gaitán-Espitia JD (2019) Beyond buying time: the role of plasticity in phenotypic adaptation to rapid environmental change. Philos Trans R Soc B: Biol Sci 374:20180174. https://doi.org/10.1098/rstb.2018.0174
Fraimout A, Jacquemart P, Villarroel B, Aponte DJ, Decamps T, Herrel A, Cornette R, Debat V (2018) Phenotypic plasticity of Drosophila suzukii wing to developmental temperature: implications for flight. J Exp Biol 221:e166868. https://doi.org/10.1242/jeb.166868
Garcia RA, Cabeza M, Rahbek C, Araújo MB (2014) Multiple dimensions of climate change and their implications for biodiversity. Science 344:1247579. https://doi.org/10.1126/science.1247579
Gleason LU, Strand EL, Hizon BJ, Dowd WW (2018) Plasticity of thermal tolerance and its relationship with growth rate in juvenile mussels (Mytilus californianus). Proc R Soc B: Biol Sci 285:20172617. https://doi.org/10.1098/rspb.2017.2617
Gracey AY, Fraser EJ, Li W, Fang Y, Taylor RR, Rogers J, Brass A, Cossins AR (2004) Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci USA 101(48):16970–16975. https://doi.org/10.1073/pnas.0403627101
Guo H et al (2017) Coastal regime shifts: rapid responses of coastal wetlands to changes in mangrove cover. Ecology 98:762–772. https://doi.org/10.1002/ecy.1698
Hansson L-A (2004) Plasticity in pigmentation induced by conflicting threats from predation and UV radiation. Ecology 85:1005–1016. https://doi.org/10.1890/02-0525
Härer A, Torres-Dowdall J, Meyer A (2017) Rapid adaptation to a novel light environment: the importance of ontogeny and phenotypic plasticity in shaping the visual system of Nicaraguan Midas cichlid fish (Amphilophus citrinellus spp.). Mol Ecol 26:5582–5593. https://doi.org/10.1111/mec.14289
Harrington RW (1961) Oviparous hermaphroditic fish with internal self-fertilization. Science 134:1749–1750. https://doi.org/10.1126/science.134.3492.1749
Harrington RW (1968) Delimitation of the thermolabile phenocritical period of sex determination and differentiation in the ontogeny of the normally hermaphroditic fish Rivulus marmoratus Poey. Physiol Zool 41:447–460
He Q, Silliman BR (2019) Climate change, human impacts, and coastal ecosystems in the anthropocene. Curr Biol 29:R1021–R1035. https://doi.org/10.1016/j.cub.2019.08.042
Helmuth BST, Hofmann GE (2001) Microhabitats, thermal heterogeneity, and patterns of physiological stress in the rocky intertidal zone. Biol Bull 201:374–384. https://doi.org/10.2307/1543615
Helmuth B et al (2016) Long-term, high frequency in situ measurements of intertidal mussel bed temperatures using biomimetic sensors. Sci Data 3:160087. https://doi.org/10.1038/sdata.2016.87
Hsu Y, Lee SP, Chen MH, Yang SY, Cheng KC (2008) Switching assessment strategy during a contest: fighting in killifish Kryptolebias marmoratus. Anim Behav 75:1641–1649. https://doi.org/10.1016/j.anbehav.2007.10.017
Huey RB, Hertz PE, Sinervo B (2003) Behavioral drive versus behavioral inertia in evolution: a null model approach. Am Nat 161:357–366. https://doi.org/10.1086/346135
Intergovernmental Panel on Climate Change (ed) (2014) Climate change 2013 – the physical science basis: working group I contribution to the fifth assessment report of the intergovernmental panel on climate change. 1st edn.Cambridge University Press. https://doi.org/10.1017/CBO9781107415324
Jones JC, Helliwell P, Beekman M, Maleszka R, Oldroyd BP (2005) The effects of rearing temperature on developmental stability and learning and memory in the honey bee, Apis mellifera. J Comp Physiol A 191:1121–1129. https://doi.org/10.1007/s00359-005-0035-z
Kanamori A et al (2016) A genetic map for the only self-fertilizing vertebrate. EG: Genes Genomes Genet 6(4):1095–1106. https://doi.org/10.1534/g3.115.022699
Kelley JL, Yee M-C, Lee C, Levandowsky E, Shah M, Harkins T, Earley RL, Bustamante CD (2012) The possibility of de novo assembly of the genome and population genomics of the mangrove rivulus, Kryptolebias marmoratus. Integr Comp Biol 52:737–742. https://doi.org/10.1093/icb/ics094
Kelley JL, Yee M-C, Brown AP, Richardson RR, Tatarenkov A, Lee CC, Harkins TT, Bustamante CD, Earley RL (2016) The genome of the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus: a model for studying phenotypic plasticity and adaptations to extreme environments. Genome Biol Evol 8:2145–2154. https://doi.org/10.1093/gbe/evw145
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Kolberg L, Raudvere U, Kuzmin I, Vilo J, Peterson H (2020) gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res9, ELIXIR-709. https://doi.org/10.12688/f1000research.24956.2
Krueger F (2015) Trim Galore!: a wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data. Babraham Institute
Langerhans RB, DeWitt TJ (2002) Plasticity constrained: over-generalized induction cues cause maladaptive phenotypes. Evol Ecol Res 4:857–870
Leriorato JC, Nakamura Y, Uy WH (2021) Cold thermal tolerance as a range-shift predictive trait: an essential link in the disparity of occurrence of tropical reef fishes in temperate waters. Mar Biol 168(6):1–18
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lieberman-Aiden E et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. https://doi.org/10.1126/science.1181369
Lin H-C, Dunson WA (1999) Phenotypic plasticity in the growth of the self-fertilizing hermaphroditic fish Rivulus marmoratus. J Fish Biol 54:250–266. https://doi.org/10.1111/j.1095-8649.1999.tb00828.x
Lins LSF, Trojahn S, Sockell A, Yee M-C, Tatarenkov A, Bustamante CD, Earley RL, Kelley JL (2018) Whole-genome sequencing reveals the extent of heterozygosity in a preferentially self-fertilizing hermaphroditic vertebrate. Genome 61:241–247. https://doi.org/10.1139/gen-2017-0188
Liu J, Frochaux M, Gardeux V, Deplancke B, Robinson-Rechavi M (2020) Inter-embryo gene expression variability recapitulates the hourglass pattern of evo-devo. BMC Biol 18:129. https://doi.org/10.1186/s12915-020-00842-z
Mackiewicz M, Tatarenkov A, Taylor DS, Turner BJ, Avise JC (2006) Extensive outcrossing and androdioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc Natl Acad Sci USA 103:9924–9928. https://doi.org/10.1073/pnas.0603847103
Mallard F, Nolte V, Schlötterer C (2020) The evolution of phenotypic plasticity in response to temperature stress. Genome Biol Evol 12:2429–2440. https://doi.org/10.1093/gbe/evaa206
Metzger DCH, Schulte PM (2018) Similarities in temperature-dependent gene expression plasticity across timescales in threespine stickleback (Gasterosteus aculeatus). Mol Ecol 27:2381–2396. https://doi.org/10.1111/mec.14591
Miller LP, Harley CDG, Denny MW (2009) The role of temperature and desiccation stress in limiting the local-scale distribution of the owl limpet, Lottia gigantea. Funct Ecol 23:756–767. https://doi.org/10.1111/j.1365-2435.2009.01567.x
Mislan KS, Wethey DS, Helmuth B (2009) When to worry about the weather: role of tidal cycle in determining patterns of risk in intertidal ecosystems. Glob Change Biol 15:3056–3065. https://doi.org/10.1111/j.1365-2486.2009.01936.x
Morris S, Taylor AC (1983) Diurnal and seasonal variation in physico-chemical conditions within intertidal rock pools. Estuar Coast Shelf Sci 17:339–355. https://doi.org/10.1016/0272-7714(83)90026-4
Mourabit S, Edenbrow M, Croft DP, Kudoh T (2011) Embryonic development of the self-fertilizing mangrove killifish Kryptolebias marmoratus. Dev Dyn 240:1694–1704. https://doi.org/10.1002/dvdy.22668
Noble DWA, Stenhouse V, Schwanz LE (2018) Developmental temperatures and phenotypic plasticity in reptiles: a systematic review and meta-analysis. Biol Rev 93:72–97. https://doi.org/10.1111/brv.12333
Oliver JC, Ramos D, Prudic KL, Monteiro A (2013) Temporal gene expression variation associated with eyespot size plasticity in Bicyclus anynana. PLoS ONE 8:e65830. https://doi.org/10.1371/journal.pone.0065830
Orizaola G, Laurila A (2008) Microgeographic variation in temperature-induced plasticity in an isolated amphibian metapopulation. Evol Ecol 23:979. https://doi.org/10.1007/s10682-008-9285-x
Peng IF, Berke BA, Zhu Y, Lee WH, Chen W, Wu CF (2007) Temperature-dependent developmental plasticity of Drosophila neurons: cell-autonomous roles of membrane excitability, CA2+ influx, and cAMP signaling. J Neurosci 27:12611–12622. https://doi.org/10.1523/JNEUROSCI.2179-07.2007
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. https://doi.org/10.1038/nbt.3122
Putnam NH et al (2016) Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res 26:342–350. https://doi.org/10.1101/gr.193474.115
R Core Team (2020) R: a language and environment for statistical computing. Vienne, Austria: R Foundation for Statistical Computing. https://www.R-project.org/. Accessed 20 April 2021.
Rhee JS, Choi BS, Kim J et al (2017) Diversity, distribution, and significance of transposable elements in the genome of the only selfing hermaphroditic vertebrate Kryptolebias marmoratus. Sci Rep 7:40121. https://doi.org/10.1038/srep40121
Riddell EA, Roback EY, Wells CE, Zamudio KR, Sears MW (2019) Thermal cues drive plasticity of desiccation resistance in montane salamanders with implications for climate change. Nat Commun 10:4091. https://doi.org/10.1038/s41467-019-11990-4
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Sawall Y, Al-Sofyani A, Hohn S, Banguera-Hinestroza E, Voolstra CR, Wahl M (2015) Extensive phenotypic plasticity of a Red Sea coral over a strong latitudinal temperature gradient suggests limited acclimatization potential to warming. Sci Rep 5:8940. https://doi.org/10.1038/srep08940
Scheel JJ (1972) Rivuline karyotypes and their evolution (Rivuline, Cyprinodontidae, Pisces). J Zool Syst Evol Res 10(1):180–209. https://doi.org/10.1111/j.1439-0469.1972.tb00797.x
Scott GR, Johnston IA (2012) Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc Natl Acad Sci USA 109(35):14247–14252. https://doi.org/10.1073/pnas.1205012109
Sentis A, Hemptinne J-L, Brodeur J (2017) Non-additive effects of simulated heat waves and predators on prey phenotype and transgenerational phenotypic plasticity. Glob Change Biol 23:4598–4608. https://doi.org/10.1111/gcb.13674
Serôdio J, Catarino F (1999) Fortnightly light and temperature variability in estuarine intertidal sediments and implications for microphytobenthos primary productivity. Aquat Ecol 33:235–241. https://doi.org/10.1023/A:1009989229098
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Tatarenkov A, Earley RL, Perlman BM, Scott Taylor D, Turner BJ, Avise JC (2015) Genetic subdivision and variation in selfing rates among Central American populations of the mangrove rivulus, Kryptolebias marmoratus. J Hered 106:276–284. https://doi.org/10.1093/jhered/esv013
Tatarenkov A, Lima SMQ, Earley RL, Berbel-Filho WM, Vermeulen FBM, Taylor DS, Marson K, Turner BJ, Avise JC (2017) Deep and concordant subdivisions in the self-fertilizing mangrove killifishes (Kryptolebias) revealed by nuclear and mtDNA markers. Biol J Lin Soc 122:558–578. https://doi.org/10.1093/biolinnean/blx103
Taylor DS (1992) Diet of the killifish Rivulus marmoratus collected from land crab burrows, with further ecological notes. Environ Biol Fish 33:389–393. https://doi.org/10.1007/BF00010951
Taylor DS (2012) Twenty-four years in the mud: what have we learned about the natural history and ecology of the mangrove rivulus, Kryptolebias marmoratus? Integr Comp Biol 52:724–736. https://doi.org/10.1093/icb/ics062
Taylor DS, Davis WP, Turner BJ (1995) Rivulus marmoratus: ecology and distributional patterns in Florida and the Central Indian River Lagoon. Bull Mar Sci 57(1):202–207
Turner BJ, Davis WP, Taylor DS (1992) Abundant males in populations of a selfing hermaphrodite fish, Rivulus marmoratus, from some Belize cays. J Fish Biol 40:307–310. https://doi.org/10.1111/j.1095-8649.1992.tb02576.x
Turner BJ, Fisher MT, Taylor DS, Davis WP, Jarrett BL (2006) Evolution of ‘maleness’ and outcrossing in a population of the self-fertilizing killifish, Kryptolebias marmoratus. Evol Ecol Res 8:1475–1486
Wang X, Bhandari RK (2020) The dynamics of DNA methylation during epigentic reprograming of primordial germ cells in medaka (Oryias latipes). Epigenetics 15(5):483–498. https://doi.org/10.1080/15592294.2019.1695341
While GM, Noble DWA, Uller T, Warner DA, Riley JL, Du W-G, Schwanz LE (2018) Patterns of developmental plasticity in response to incubation temperature in reptiles. J Exp Zool Part A: Ecol Integr Physiol 329:162–176. https://doi.org/10.1002/jez.2181
Williams CM, Henry HAL, Sinclair BJ (2015) Cold truths: how winter drives responses of terrestrial organisms to climate change. Biol Rev 90(1):214–235
Wolfe K, Nguyen HD, Davey M, Byrne M (2020) Characterizing biogeochemical fluctuations in a world of extremes: a synthesis for temperate intertidal habitats in the face of global change. Glob Change Biol 26:3858–3879. https://doi.org/10.1111/gcb.15103
Xu S, Hu E, Cai Y, Xie Z, Luo X, Zhan L, Tang W, Wang Q, Liu B, Wang R, Xie W, Wu T, Xie L, Yu G (2024) Using clusterProfiler to characterize multiomics data. Nat Protoc 19(11):3292–3320. https://doi.org/10.1038/s41596-024-01020-z
Zhu Q, Zhang L, Li L, Que H, Zhang G (2016) Expression characterization of stress genes under high and low temperature stresses in the Pacific oyster, Crassostrea gigas. Mar Biotechnol 18:176–188. https://doi.org/10.1007/s10126-015-9678-0
Acknowledgements
The research was approved by the Institutional Animal Care and Use Committee at the University of Alabama, Protocol #16-09-0133. This research used resources of the Center for Institutional Research Computing at Washington State University. We would like to acknowledge the University of Alabama Honors College, the University of Alabama Randall Research Scholars Program.
Funding
This work was supported by the FNRS – Crédit De Recherche (J.0189.20; ‘Epigenetic diversity origin in Rivulus’; BE to FS), a Grant-in-Aid for Scientific Research from JSPS (19 K06738 to AK), and a Sigma Xi Grants-in-Aid (to CC).
Author information
Authors and Affiliations
Contributions
□Anthony A. Snead o Data Curation o Formal analysis o Methodology o Visualization o Writing – original draft o Writing – review & editing o Software □ Corey R. Quackenbush o Investigation o Methodology □ Shawn Trojahn o Investigation o Methodology □ Anna L. McDonald o Investigation o Methodology o Software o Writing – review & editing □ Luana Lins o Conceptualization o Data Curation o Methodology o Writing – review & editing □ Chris Cornelius o Investigation □ Paula E Adams o Conceptualization o Investigation □ Dengke Ma o Conceptualization o Funding acquisition □ Yuying Hsu o Conceptualization o Writing – review & editing o Funding acquisition □ Eric Haag o Conceptualization o Funding acquisition □ Frédéric Silvestre o Conceptualization o Writing – review & editing o Funding acquisition □ Akira Kanamori o Conceptualization o Writing – review & editing o Funding acquisition □ Ryan L Earley o Conceptualization o Project administration o Methodology o Resources o Supervision o Funding acquisition □ Joanna L. Kelley o Conceptualization o Methodology o Project Administration o Resources o Supervision o Visualization o Writing – review & editing o Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Snead, A.A., Quackenbush, C.R., Trojahn, S. et al. Embryonic thermal environments drive plasticity in gene expression. Fish Physiol Biochem 51, 111 (2025). https://doi.org/10.1007/s10695-025-01522-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10695-025-01522-x