Abstract
Lung cancer remains the most prevalent carcinoma with a high mortality rate, yet the underlying mechanisms driving pulmonary neoplasia and disease progression are not fully understood. In our study, we conducted a comprehensive analysis of the transcriptome profiles and clinicopathological characteristics of 515 patients diagnosed with non-small cell lung cancer (NSCLC) from the TCGA database. We identified a significant upregulation of centromere protein M (CENPM) in NSCLC tissues, which was positively correlated with poor prognosis. Furthermore, overexpression of CENPM markedly promoted cell proliferation and increased the tumorigenic potential of NSCLC cell lines (A549/NCI-H1299), leading to accelerated tumor progression and reduced survival time in tumor-bearing mice. Mechanistically, CENPM activated the Wnt/β-catenin signaling pathway via the cell division cycle 20 (CDC20)/MYB proto-oncogene-like 2 (MYBL2) axis. Inhibition of either Wnt signaling or the CDC20/MYBL2 axis attenuated the tumorigenic potential and proliferative effects induced by CENPM. Our findings underscore the critical role of CENPM in driving NSCLC development and suggest that CENPM could serve as a novel biomarker for predicting NSCLC progression in clinical settings.
Graphical abstract
CENPM stimulates the expression of the cell differentiation cycle protein CDC20, leading to the upregulation of MYBL2. Consequently, the CDC20/MYBL2 axis activates the pro-survival Wnt/β-catenin signaling pathway, enhancing the tumorigenic potential and proliferative characteristics of lung cancer cells. This study underscores the crucial role of CENPM in promoting NSCLC development through the CDC20/MYBL2 signaling pathway. Thus, CENPM may serve as a novel biomarker for predicting NSCLC progression in clinical settings.

Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with approximately 2.2 million new cases diagnosed and 1.8 million deaths in 2020, accounting for nearly one-fifth (18.0%) of all cancer deaths (Rosell and Karachaliou 2015; de Groot et al. 2018; Thandra et al. 2021). NSCLC constitutes 85% of all lung carcinomas (Herbst et al. 2018). Despite significant advances in diagnosis and therapeutic approaches, NSCLC continues to have one of the poorest prognoses among cancers. Emerging research has focused on identifying actionable molecular alterations, leading to the ongoing evolution of targeted treatments. However, the majority of NSCLC patients still experience tumor recurrence or distant metastasis within five years of diagnosis, with a five-year overall survival rate of just 18.2% (Rotow and Bivona 2017). Therefore, identifying innovative prognostic biomarkers and gaining a better understanding of the mechanisms underlying tumor progression are crucial for improving clinical diagnosis and developing novel therapeutic strategies for NSCLC.
CENPM, also named as proliferation-associated nuclear element 1 (PANE1), is a component of CENPA nucleosome‑associated complex (NAC) (Qi et al. 2022; Liu et al. 2020). It plays a vital role in the assembly of kinetochore proteins, chromosome segregation, and mitotic progression. Aberrant function of CENPM can lead to chromosomal instability (CIN) (McKinley and Cheeseman 2016), a hallmark of cancer that contributes to aneuploidy and tumorigenesis (Pino and Chung 2010; Qi et al. 2022). Moreover, the proliferative characteristics of cancer cells is tightly correlated with mitosis and chromosome separation (Wassmann and Benezra 2001). Indeed, numerous studies have demonstrated that abnormal expression of the CENPA complex is observed in several tumor types. For instance, upregulation of CENPA has been shown to stimulate tumor progression in colorectal cancer (Tomonaga et al. 2003), and increasing evidence suggested that CENPF contributed to a poor prognosis in patients with breast cancer and nasopharyngeal carcinoma (Sun et al. 2016). However, the specific mechanisms by which the CENPA complex contributes to tumor progression remain unclear. Recently, CENPM has been investigated in various cancer types and has emerged as a novel prognostic marker in liver cancer, melanoma, breast cancer, and bladder cancer (Xiao et al. 2019; Liu et al. 2020; Tong et al. 2024). Despite these findings, the relationship between CENPM and NSCLC progression has been relatively unexplored. While some studies suggest that the CENPM gene may play a role in the development of lung adenocarcinoma (Zhou et al. 2021), its specific function in NSCLC progression remains to be fully elucidated.
Our study conducted an initial screening of CENPM transcriptome expression in 515 NSCLC patients, suggesting the clinical relevance of CENPM in patient samples, demonstrating that its expression was closely associated with distant metastasis and reduced survival time in NSCLC patients. Mechanistically, CENPM overexpression significantly enhanced NSCLC cell proliferation and tumorigenic potential both in vitro and in vivo through the activation of Wnt/β-catenin signaling via the CDC20/MYBL2 axis. Thus, our study underscores the critical role of CENPM in NSCLC development and suggests its potential as a novel prognostic biomarker and therapeutic target for NSCLC patients.
Materials and methods
Cell lines and chemical reagents
Human NSCLC cell lines NCI-H1299 and A549 were purchased from the Global Bioresource Center (ATCC, USA). CENPM overexpression A549/NCI-H1299 cells were established by Tsingke (China) and determined by western blotting. Cancer cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher, USA) containing 10% fetal calf serum (FBS) in a standard atmosphere with 5% CO2, and at a temperature of 37 °C. cAMP inhibitor PKI 14–22 amide, myristoylated (PKI), Wnt/β-catenin inhibitor PNU-74,654 (PUN), hedgehog signaling inhibitor KAAD-Cyclopamine (KAAD) were purchased from Medchemexpress (USA).
Clinical specimens
Information of 59 normal tissues and 721 NSCLC tissues in the Cancer Genome Atlas Program (TCGA) databases were downloaded from Cbioportal database and http://ualcan.path.uab.edu/index.html. 211 NSCLC patients (containing transcription profile information) were divided into high-CENPM and low-CENPM groups according to the mean expression of CENPM. Paraffin sections of NSCLC tissues were obtained from Affiliated Central Hospital of Dalian University of Technology. 26 patients were divided into metastatic (M1) and non-metastatic (M0) groups according to their post-treatment diagnosis (five-years). Patients were informed with written consent and agreed to take part in our clinical experiments. All experiments were conducted according to the Declaration of Helsinki. Ethical review was granted by the Ethics Committee of the Affiliated Central Hospital of Dalian University of Technology.
Cell proliferation assay
Cell proliferation was determined by a cell counting kit (CCK8) purchased from Solarbio (China). 3000 pre-treated cells were seeded in 96-well plates. The absorbance value was measured at 0, 24, 48 and 72 h after CCK8 reagent incubation according to the manufacturer’s instructions. Each experiment was conducted for at least three independent times.
Colony formation assay
For tumorigenic potential assay, pre-treated cells were seeded in 6-well plates (500 cells/ well). Then cells were cultured with serum-free culture medium. After 10 days, cell colony formation was observed under the inverted optical microscope (Leica, Germany). Meanwhile, colonies were washed, and stained with crystal violet. The visible colonies were counted and analyzed. Each experiment was conducted for at least three independent times.
Cell apoptosis assay
Cell apoptosis was evaluated using an Annexin V/PI staining kit (Solarbio, China) and flow cytometry analysis. Briefly, siRNA treated tumor cells were harvested and then stained by Annexin V/PI staining kit according to the manufacturer’s protocol. Then cell apoptosis was determined by flow cytometry (BD Pharmingen, CA). Each experiment was conducted for at least three independent times.
Small interfering RNA (siRNA)
SiRNA for CDC20, CENPM and MYBL2 was purchased from Tsingke (China). The sequences of the CENPM siRNAs were as follows: 5′-GGACUCGAUGCUCAAAGAGTT-3′ and 5′-CUGAAGGUCCACUUGGCAATT-3′; The sequences of the CDC20 siRNAs were as follows: 5′-GGAGCUCAUCUCAGGCCA UUU-3′ and 5′- GGGAAUAUAUAUCCUCUGUUU-3′; The sequences of the MYBL2 siRNAs were as follows: 5′- GAGGGATAGCAAGTGCAAGGT-3′ and 5′-TTCCAGTCCTGCTGTCCAAA-3′; SiRNA interference was conducted using an Lipofectamine 3000 (Invitrogen, USA) protocol. The siRNA was added at the of concentration 100 nmol/l. Each experiment was conducted for at least three independent times.
Western blotting
RIPA lysis buffer (Abcam, UK) was used for whole cell lysates extraction. Total proteins were separated by 10% SDS-PAGE, and samples were then transferred onto a nitrocellulose membrane (Thermo, USA). Subsequently, samples were incubated with the primary antibodies at 4 °C overnight. After wash with TBST, samples were incubated with a horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Primary antibodies anti-β-catenin (ab32572), anti-CENPM (ab243820), anti-CDC20 (ab26483), anti-MYBL2 (ab191064) and anti-Wnt3a (ab219412) were purchased from Abcam (UK). The protein quantification was performed by Image J 5.0 software. Each experiment was conducted for at least three independent times.
Real-time quantitative polymerase chain reaction (PCR)
Total RNA from vector or CENPM overexpressed tumor cells was extracted using Trizol reagent (Thermo Fisher, USA) according to the manufacturer’s protocol. Reverse transcription was performed using the SuperScript II RNase H-RT kit (Thermo Fisher, USA) with 1 µg of RNA from each sample to generate cDNA. All primers were designed according to the guidance of PrimerBank. Real-time PCR was conducted in a 20 µL reaction system containing primers, cDNA, SYBR master mix, and ddH₂O. GAPDH was used as the internal control. Gene expression changes were analyzed using the 2-∆∆CT method. The sequences of primers were list as follow:
Gene | Sequence of primers |
|---|---|
CENPM (forward) | 5′-GCGGACTCGATGCTCAAAGA-3′ |
CENPM (reverse) | 5′-TTCTGGAGACTGTATTTGCTGTG-3′ |
CDC20 (forward) | 5′-GCACAGTTCGCGTTCGAGA-3′ |
CDC20 (reverse) | 5′-CTGGATTTGCCAGGAGTTCGG-3′ |
MYBL2 (forward) | 5′-CCGGAGCAGAGGGATAGCA-3′ |
MYBL2 (reverse) | 5′-CAGTGCGGTTAGGGAAGTGG-3′ |
GAPDH (forward) | 5′-TGTGGGCATCAATGGATTTGG-3′ |
GAPDH (reverse) | 5′-ACACCATGTATTCCGGGTCAAT-3′ |
Immunostaining
Sections from NSCLC patients or mice were blocked with 5% bovine serum albumin for 30 min, then incubated with following primary antibodies anti-CENPM (ab243820, Abcam, UK) anti-Wnt3a (ab219412, Abcam, UK), anti-β-catenin (ab32572, Abcam, UK) and anti-Ki67 (ab15580, Abcam, UK) overnight at 4℃. Subsequently, sections were incubated with secondary antibody for 2 h and developed at room temperature, according to the manufacturer’s instructions. Protein expression intensity was calculated by Image-Pro Plus 5.0 (Media cybernetics, USA).
Animal protocols
NOD-SCID mice (female, 6 ~ 8 weeks) were obtained from Vitalriver (USA) and raised in a specific pathogen free (SPF) room. For tumor growth assay in vivo, Immune deficient mice were subcutaneously injected with 1 × 106 A549 cells (6 mice per group). Tumor tissues were collected for further protein expression assay. Tumor volume was calculated by the formula: tumor volume = length × width 2/2. Animal experiments were approved and monitored by the Animal Ethics Committee of Affiliated Central Hospital of Dalian University of Technology. The animal experiments were performed in accordance with the Public Health Service Policy and the WHO guidelines for the humane use and care of animals.
Statistical analysis
All data in our study are presented as the mean values ± SD. Each experiment was conducted for at least three independent times. Variation analysis was performed using one-way ANOVA or t test. Multiple comparisons among the groups were analyzed by Sidak correction method. The survival rates were determined by Kaplan–Meier survival analysis. Statistical analysis was conducted by SPSS 23.0 software (IBM, USA) or GraphPad Prism version 6.0 (GraphPad Software, USA). P-values < 0.05 were considered as a statistically significant difference.
Results
Upregulated CENPM in NSCLC tissues is associated with poor prognosis
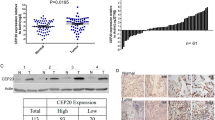
To evaluate the clinical relevance of CENPM in NSCLC development, we analyzed the transcriptome expression of CENPM in 515 NSCLC patients and 59 normal tissues using data from the TCGA database. The results showed that CENPM expression was significantly upregulated in tumor tissues compared to normal tissues, with no significant differences observed across different clinical stages of NSCLC (Fig. 1a). Given the high expression of CENPM in NSCLC tissues, we next examined its impact on overall survival. We categorized 211 NSCLC patients into high-CENPM (n = 103) and low-CENPM (n = 108) expression groups. Patients with lower CENPM expression had significantly longer survival times compared to those with high expression (Fig. 1b), suggesting that high CENPM levels are associated with poor prognosis in NSCLC. To further explore the role of CENPM in NSCLC metastasis, we analyzed 26 NSCLC tissue samples from clinical patients, categorized into metastatic (M1) and non-metastatic (M0) groups based on post-treatment diagnoses over a five-year period. Immunohistochemical analysis of these tissues revealed that CENPM expression was significantly higher in metastatic patients (Fig. 1c). These findings provide evidence that CENPM upregulation contributes to tumorigenesis and is linked to poor prognosis in NSCLC patients.
CENPM promoted cell proliferation and tumorigenic potential in NSCLC
Given the association between elevated CENPM expression and poor prognosis in NSCLC patients, we further investigated the molecular mechanisms underlying CENPM-driven NSCLC progression. NSCLC cell lines NCI-H1299 and A549 were treated with CENPM siRNA and cultured for three days (Fig. 1d). Silencing of CENPM in A549 and NCI-H1299 cells resulted in increased apoptosis (Fig. 1e), underscoring the critical role of CENPM in regulating NSCLC cell survival. To further explore the impact of CENPM on NSCLC progression, we established CENPM-overexpressing A549 and NCI-H1299 cells (Fig. 1f) and evaluated their proliferation and colony-forming abilities. Notably, CENPM overexpression significantly increased cell proliferation compared to control cells (Fig. 1g). Additionally, CENPM-overexpressing cells showed enhanced colony formation capacity (Fig. 1h). These findings indicate that elevated CENPM expression promotes NSCLC cell proliferation and tumorigenic potential.
CENPM promoted NSCLC progression and correlated with poor prognosis a mRNA expression of CENPM in normal tissues (n = 59) and NSCLC tissues (n = 277 in stage I, n = 125 in stage II, n = 85 in stage III and n = 28 in stage IV) from TCGA database. b Overall survival curve of 211 NSCLC patients divided into high CENPM (n = 103) and low CENPM (n = 108) groups. c Immunohistochemistry of CENPM in NSCLC divided into metastatic (M1, n = 13) and non-metastatic (M0, n = 13) groups. The scale bar was 100 μm. Expression intensity were quantified in 26 patients. d Relative CENPM expression of A549 and NCI-H1299 cells treated with scrambled and CENPM siRNA, determined by qPCR. e Cell apoptosis of A549 and NCI-H1299 cells treated with scrambled and siRNA. f Western blotting of CENPM in vector and CENPM overexpression A549/NCI-H1299 cells. g Cell proliferation of vector and CENPM overexpression A549/NCI-H1299 cells. h Colony formation of vector and CENPM overexpression A549/NCI-H1299 cells
CENPM promoted NSCLC development through Wnt/β-catenin signaling
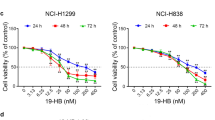
To elucidate the molecular pathways through which CENPM promotes NSCLC development, we stratified 211 NSCLC patients into high-CENPM (n = 126) and low-CENPM (n = 85) groups according to the mean expression of CENPM using data from the TCGA database. Enrichment analysis identified differentially expressed genes and key pathways associated with CENPM (Fig. 2a). Notably, the cAMP, Wnt, and Hedgehog pro-survival signaling pathways were highlighted, as they play critical roles in tumor progression. To investigate these pathways, we treated vector and CENPM-overexpressing A549 and NCI-H1299 cells with a cAMP inhibitor (PKI), a Wnt/β-catenin inhibitor (PNU), and a Hedgehog signaling inhibitor (KAAD). Blockade of the Wnt/β-catenin pathway significantly inhibited both proliferation (Fig. 2b) and tumorigenic potential (Fig. 2c) in CENPM-overexpressing cells, while PKI and KAAD treatments had minimal effects. These findings suggest that CENPM mediates NSCLC progression in a Wnt/β-catenin-dependent manner. Our preliminary experiments further supported this hypothesis: Wnt-3a mRNA levels were upregulated in CENPM-overexpressing A549 cells, with minimal changes observed for WNT1 and WNT2 (not shown in data). To validate these findings, we conducted Western blot analysis to assess Wnt-3a and β-catenin expression in CENPM-overexpressing A549 and NCI-H1299 cells. Consistent with our hypothesis, both Wnt-3a and β-catenin expression were elevated in these cells compared to vector controls (Fig. 2d). Additionally, immunostaining of 26 tumor tissues from metastatic NSCLC patients confirmed that Wnt-3a was upregulated in these samples (Fig. 2e). Collectively, our data suggest that CENPM promotes NSCLC development through the activation of Wnt/β-catenin signaling.
CENPM promoted NSCLC development through Wnt/β-catenin signaling a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in CENPM high (n = 126, relative CENPM expression > 100) in comparison with low CENPM (n = 85, relative CENPM expression < 100) NSCLC patients. b Cell proliferation of vector A549/NCI-H1299, CENPM overexpression A549/NCI-H1299 treated with PBS, PKI (20 nM), PNU (10 nM) and KAAD (10 nM) respectively. c Colony formation of vector A549/NCI-H1299, CENPM overexpression A549/NCI-H1299 treated with PBS, PKI (20 nM), PNU (10 nM) and KAAD (10 nM) respectively. d Western blotting of Wnt-3a and β-catenin in vector and CENPM overexpression A549/NCI-H1299. e Immunofluorescence of Wnt-3a in NSCLC tissues divided into metastatic (M1, n = 13) and non-metastatic (M0, n = 13) groups. The scale bar was 50 μm. Expression intensity were quantified in 26 patients
CENPM upregulated CDC20 and MYBL2 to mediate Wnt/β-catenin signaling activation
Given the role of Wnt/β-catenin signaling in CENPM-induced tumor progression, we sought to investigate how CENPM mediates Wnt signaling activation. We further analyzed the genes upregulated in CENPM-high patients compared to CENPM-low patients (n = 211). The upregulated genes in the CENPM-high group are listed in Fig. 3a. Notably, CDC20 and MYBL2 were significantly upregulated in the CENPM-high group. Previous studies have shown that the MYBL2/CDC20 axis can stimulate gastric cancer cell proliferation through Wnt/β-catenin signaling (Deng et al. 2021). To examine whether CENPM influences CDC20 and MYBL2 expression in NSCLC, we assessed the levels of these genes in CENPM-overexpressing A549 and NCI-H1299 cells. The results showed increased expression of both CDC20 and MYBL2 (Fig. 3b), suggesting that CENPM upregulates these genes in NSCLC cells. Next, we investigated whether CENPM mediates Wnt/β-catenin signaling activation via the CDC20/MYBL2 axis. Using siRNA technology, we silenced CDC20 and MYBL2 in CENPM-overexpressing A549 and NCI-H1299 cells (Fig. 3c and d). Western blot analysis was then performed to assess Wnt-3a and β-catenin expression in CDC20- or MYBL2-silenced cells. Consistent with our hypothesis, silencing CDC20 or MYBL2 suppressed the upregulation of Wnt-3a and β-catenin in CENPM-overexpressing cells (Fig. 3e and f).Furthermore, silencing CDC20 or MYBL2 reduced cell proliferation (Fig. 3g and h) and tumorigenic potential (Fig. 3i and j) induced by CENPM. These results suggest that CENPM upregulates CDC20 and MYBL2 to activate Wnt/β-catenin signaling in NSCLC cells.
CENPM upregulated CDC20 and MYBL2 to mediate Wnt/β-catenin signaling activation a heatmap of top 30 upregulated genes in high CENPM patients (n = 126, relative CENPM expression > 100) in comparison with low CENPM patients (n = 85, relative CENPM expression < 100). b Western blotting of CDC20 and MYBL2 in vector and CENPM overexpression A549/NCI-H1299 cells. c Relative expression of CDC20 in CENPM overexpression A549/NCI-H1299 treated with scrambled and CDC20 siRNA, determined by qPCR. d Relative expression of MYBL2 in CENPM overexpression A549/NCI-H1299 treated with scrambled and MYBL2 siRNA, determined by qPCR. e Western blotting of Wnt-3a and β-catenin in CENPM overexpression A549/NCI-H1299 treated with scrambled and CDC20 siRNA. f Western blotting of Wnt-3a and β-catenin in CENPM overexpression A549/NCI-H1299 treated with scrambled and MYBL2 siRNA. g Cell proliferation of A549/NCI-H1299 treated with scrambled and CDC20 siRNA. h Cell proliferation of A549/NCI-H1299 treated with scrambled and MYBL2 siRNA. i Colony formation of A549/NCI-H1299 treated with scrambled and CDC20 siRNA. j Colony formation of A549/NCI-H1299 treated with scrambled and MYBL2 siRNA
CENPM promoted NSCLC progression in vivo
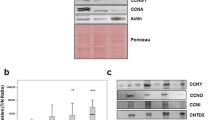
To assess the pro-tumor effects of CENPM in vivo, we evaluated the tumor growth of CENPM-overexpressing cells in immunodeficient mice. A total of 1 × 106 vector or CENPM-overexpressing A549 cells were subcutaneously implanted into the mice. As expected, CENPM-overexpressing A549 cells formed rapidly growing tumors compared to the vector group (Fig. 4a). Additionally, mice bearing CENPM-overexpressing A549 tumors exhibited significantly shorter survival times (Fig. 4b). Consistent with our in vitro findings, the protein levels of CDC20, MYBL2, Wnt-3a, and β-catenin were upregulated in CENPM-overexpressing tumor tissues compared to the vector group (Fig. 4c and d). We also observed a marked increase in the expression of Ki67, a cell proliferation marker, in CENPM-overexpressing tumor tissues (Fig. 4e). Collectively, these results suggest that CENPM promotes NSCLC progression by regulating the CDC20/MYBL2/Wnt signaling pathway.
CENPM promoted NSCLC progression in vivo a tumor volume of vector or CENPM overexpression A549 bearing mice. b Overall survival of vector or CENPM overexpression A549 bearing mice. c Western blotting of CDC20 and MYBL2 in tumor tissues isolated from vector or CENPM overexpression A549 bearing mice. d Western blotting of Wnt-3a and β-catenin in tumor tissues isolated from vector or CENPM overexpression A549 bearing mice. e Immunofluorescence of Ki67 in tumor tissues isolated from vector or CENPM overexpression A549 bearing mice
Discussion
CENPM is not only a microtubule-associated kinetochore protein involved in chromosome segregation but also plays a role in regulating the cell cycle (Earnshaw 2015). It has been reported that elevated CENPM expression can lead to uneven chromosome segregation, resulting in an unequal distribution of chromosomes. These aneuploid cells are prone to chromosomal aberrations and display more visible abnormalities than normal cells (Dobie et al. 1999; Yang et al. 2022). Consistently, CENPM is upregulated in proliferating cells, such as activated lymphoid and cancer cells (Earnshaw 2015). Previous studies have shown that CENPM expression is strongly associated with the prognosis of patients with hepatocellular carcinoma and can complement AFP in diagnosing liver cancer (Z. H. Wu and Yang 2020). Moreover, recent investigations addressed that CENPM might contribute to tumor metastasis and recurrence in melanoma (Chen et al. 2019), bladder cancer (Kim et al. 2018) and pancreatic cancer (Zheng et al. 2020). However, limited research has explored the role of CENPM in the oncogenesis of NSCLC. Our study systematically analyzed the transcriptome profiles and clinicopathological characteristics of 515 NSCLC patients from the TCGA dataset, revealing that CENPM is upregulated in tumor tissues and closely associated with distant metastasis and poor survival in NSCLC patients. These findings suggest that CENPM may promote NSCLC progression and be linked to tumor cell proliferation and invasiveness. Further, our investigation demonstrated that depletion of CENPM inhibited cell proliferation and tumorigenic potential in vitro. In addition, we established a CENPM-overexpressing xenograft mouse model to show that CENPM plays a critical role in tumor growth and disease progression in vivo. Taken together, these findings imply that CENPM could serve as a novel biomarker for early diagnosis or prognosis of NSCLC and present a potential chemotherapeutic target for NSCLC treatment.
Given CENPM’s central role in NSCLC development, we further explored the molecular mechanisms by which CENPM promotes cell proliferation and tumorigenesis. Previous studies have reported that CENPM regulates cell proliferation and invasion through the mTOR/p70S6K axis in pancreatic cancer (Zheng et al. 2020). Additionally, CENPM has been shown to suppress apoptosis and promote cell cycle progression via p53 signaling in hepatocarcinogenesis (Xiao et al. 2019). Notably, a study on the protein interaction network of CENPM demonstrated that it mediates tumor development by modulating cell cycle-associated proteins such as KIF4A, HMMR, CKS2, and CDC25C key drivers of carcinogenesis (Xiao et al. 2019). Consistent with these reports, our study found significant upregulation of CDC20 and MYBL2 in CENPM-overexpressing NSCLC cell lines and tumor tissues. Both CDC20 and MYBL2 are well-recognized as central regulators of cell proliferation and differentiation involved in tumorigenesis. CDC20, a key component of the spindle assembly checkpoint, is associated with cell cycle regulation, proliferation, and apoptosis (Yu 2002; Jeong et al. 2022). Its overexpression enhances cell proliferation and migratory capacity and plays a vital role in tumorigenesis and progression in various cancers (Deng et al. 2021). Similarly, evidence suggests that upregulated MYBL2, alongside CDC20, can serve as a biomarker for disease severity in several cancers, including glioma, gastric cancer, lung cancer, and breast cancer (Wang et al. 2015; Musa et al. 2017). Recent research also revealed that MYBL2 and FOXM1 regulate a crucial “driver network” consisting of 26 genes, including CDC20, that are involved in cell cycle modulation and proliferation in lung cancer (Ahmed 2019). However, until now, these findings were primarily based on gene expression network analysis, and the specific underlying molecular mechanisms remained largely unclear. For the first time, our experiments demonstrated that CENPM-mediated upregulation of CDC20 and MYBL2 could further activate downstream Wnt/β-catenin signaling in NSCLC.
The Wnt/β-catenin signaling pathway plays a crucial role in various biological processes, from embryogenesis to growth-related diseases (MacDonald et al. 2009). Aberrant activation of this pathway is a key driver of tumor replase and growth in various cancers (Zhao et al. 2022; Fodde and Brabletz 2007). The pro-tumorigenic role of Wnt/β-catenin signaling has been extensively studied in recent years. For example, Teng et al. demonstrated that activation of the Wnt/β-catenin pathway enhances stem-like properties in lung cancer, such as increased colony formation, tumorigenicity, and drug resistance (Teng et al. 2010). Furthermore, numerous studies have shown that upregulation of the Wnt/β-catenin axis promotes epithelial-mesenchymal transition, migration, and invasiveness of cancer cells through various mechanisms. While β-catenin mutations contribute to aberrant activation of Wnt signaling in certain cancers, such as gastric and colon cancers, these mutations are rarely detected in NSCLC (Wu et al. 2012). Therefore, identifying regulators of Wnt signaling could provide new insights into the molecular mechanisms underlying NSCLC. Our study demonstrated that CENPM upregulates CDC20 and MYBL2, which in turn significantly increases Wnt-3a and β-catenin expression, providing a novel mechanism supporting the pro-tumorigenic role of Wnt/β-catenin signaling in lung cancer. Notably, conductin (also known as axin2/axil), a negative regulator of Wnt/β-catenin signaling that localizes and functions at centrosomes and the mitotic apparatus, can be degraded by CDC20 (Hadjihannas et al. 2012). This suggests that CDC20 may activate the Wnt/β-catenin axis by promoting conductin degradation. Our findings highlight the importance of CDC20- and MYBL2-mediated activation of Wnt/β-catenin signaling in NSCLC progression, offering a direction for future research. Moreover, discovering and validating small-molecule inhibitors targeting CDC20 or MYBL2 could represent a promising therapeutic strategy for NSCLC.
Given the limitations of previous studies, our research identified the significant clinical relevance of CENPM in NSCLC by analyzing transcriptomic profiles and clinical characteristics from the TCGA database, as well as pathological sections from clinical samples. Initially, we confirmed the crucial role of CENPM in NSCLC. Subsequently, we established CENPM-overexpressing cell lines, along with CDC20 and MYBL2 knockdown cell lines, to further investigate the underlying molecular mechanisms. Our findings demonstrated that overexpression of CENPM leads to reduced apoptosis and increased cell proliferation and tumorigenesis in NSCLC through CDC20/MYBL2-mediated activation of the Wnt/β-catenin signaling pathway. Notably, our CENPM-overexpressing NSCLC mouse model further validated this pro-tumor mechanism in vivo. Our study suggests that CENPM and its associated pathways could serve as potential prognostic markers and therapeutic targets in NSCLC. However, there are several limitations to our study. The patient sample size was relatively small, and the number of pathological sections analyzed was limited, which constrains the statistical power of our findings. Nonetheless, our preliminary results provide valuable insights for ongoing research. Furthermore, the synergistic interactions and precise molecular mechanisms between CDC20 and MYBL2 require further exploration. Additional details of the signaling pathway, such as the roles of other proteins like TCF4 in the Wnt pathway, also need to be investigated.
In conclusion, our research indicates that CENPM promotes NSCLC progression via CDC20/MYBL2/Wnt signaling and provides a promising biomarker for the individualized treatment of NSCLC patients.
Data availability
The article contains all the data in the study.
Abbreviations
- PKI:
-
14–22 amide, myristoylated
- PUN:
-
PNU-74654
- KAAD:
-
KAAD-Cyclopamine
- NSCLC:
-
Non-small cell lung cancer
- TCGA:
-
The Cancer Genome Atlas Program
- CENPM:
-
Centromere Protein M
- CDC20:
-
Cell Division Cycle 20
- MYBL2:
-
MYB Proto-Oncogene Like 2
- CIN:
-
Chromosomal instability
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PANE1:
-
Proliferation associated nuclear element 1
- NAC:
-
Nucleosome‑associated complex
References
Ahmed F (2019) Integrated network analysis reveals FOXM1 and MYBL2 as key regulators of cell proliferation in non-small cell lung cancer. Front Oncol 9:1011. https://doi.org/10.3389/fonc.2019.01011
Chen J, Wu F, Shi Y, Yang D, Xu M, Lai Y et al (2019) Identification of key candidate genes involved in melanoma metastasis. Mol Med Rep 20(2):903–914. https://doi.org/10.3892/mmr.2019.10314
de Groot PM, Wu CC, Carter BW, Munden RF (2018) The epidemiology of lung cancer. Transl Lung Cancer Res 7(3):220–233. https://doi.org/10.21037/tlcr.2018.05.06
Deng Q, Wu L, Li Y, Zou L (2021) MYBL2 in synergy with CDC20 promotes the proliferation and inhibits apoptosis of gastric cancer cells. Adv Clin Exp Med 30(9):957–966. https://doi.org/10.17219/acem/135938
Dobie KW, Hari KL, Maggert KA, Karpen GH (1999) Centromere proteins and chromosome inheritance: a complex affair. Curr Opin Genet Dev 9(2):206–217. https://doi.org/10.1016/s0959-437x(99)80031-8
Earnshaw WC (2015) Discovering centromere proteins: from cold white hands to the A, B, C of CENPs. Nat Rev Mol Cell Biol 16(7):443–449. https://doi.org/10.1038/nrm4001
Fodde R, Brabletz T (2007) Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol 19(2):150–158. https://doi.org/10.1016/j.ceb.2007.02.007
Hadjihannas MV, Bernkopf DB, Brückner M, Behrens J (2012) Cell cycle control of Wnt/β-catenin signalling by conductin/axin2 through CDC20. EMBO Rep 13(4):347–354. https://doi.org/10.1038/embor.2012.12
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553(7689):446–454. https://doi.org/10.1038/nature25183
Jeong SM, Bui QT, Kwak M, Lee JY, Lee PC (2022) Targeting Cdc20 for cancer therapy. Biochim Biophys Acta Rev Cancer 1877(6):188824. https://doi.org/10.1016/j.bbcan.2022.188824
Kim WT, Seo SP, Byun YJ, Kang HW, Kim YJ, Lee SC et al (2018) The anticancer effects of garlic extracts on bladder cancer compared to cisplatin: a common mechanism of action via centromere protein M. Am J Chin Med 46(3):689–705. https://doi.org/10.1142/s0192415x18500362
Liu Y, Yu W, Ren P, Zhang T (2020) Upregulation of centromere protein M promotes tumorigenesis: a potential predictive target for cancer in humans. Mol Med Rep 22(5):3922–3934. https://doi.org/10.3892/mmr.2020.11461
MacDonald BT, Tamai K, He X (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17(1):9–26. https://doi.org/10.1016/j.devcel.2009.06.016
McKinley KL, Cheeseman IM (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17(1):16–29. https://doi.org/10.1038/nrm.2015.5
Musa J, Aynaud MM, Mirabeau O, Delattre O, Grünewald TG (2017) MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 8(6):e2895. https://doi.org/10.1038/cddis.2017.244
Pino MS, Chung DC (2010) The chromosomal instability pathway in colon cancer. Gastroenterology 138(6):2059–2072. https://doi.org/10.1053/j.gastro.2009.12.065
Qi N, Niu Y, Li Z, Xiao L, Tang D, Gao W (2022) The prognostic value and mechanisms of centromere protein M in patients with lung adenocarcinoma. Transl Cancer Res 11(10):3471–3490. https://doi.org/10.21037/tcr-22-491
Rosell R, Karachaliou N (2015) Lung cancer in 2014: optimizing lung cancer treatment approaches. Nat Rev Clin Oncol 12(2):75–76. https://doi.org/10.1038/nrclinonc.2014.225
Rotow J, Bivona TG (2017) Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 17(11):637–658. https://doi.org/10.1038/nrc.2017.84
Sun X, Clermont PL, Jiao W, Helgason CD, Gout PW, Wang Y et al (2016) Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer 139(4):899–907. https://doi.org/10.1002/ijc.30133
Teng Y, Wang X, Wang Y, Ma D (2010) Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Biophys Res Commun 392(3):373–379. https://doi.org/10.1016/j.bbrc.2010.01.028
Thandra KC, Barsouk A, Saginala K, Aluru JS, Barsouk A (2021) Epidemiology of lung cancer. Contemp Oncol (Pozn) 25(1):45–52. https://doi.org/10.5114/wo.2021.103829
Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T et al (2003) Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res 63(13):3511–3516
Tong Y, Zhou T, Wang X, Deng S, Qin L (2024) Upregulation of CENPM promotes breast carcinogenesis by altering immune infiltration. BMC Cancer 24(1):54. https://doi.org/10.1186/s12885-023-11808-z
Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W (2015) Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther 151:141–151. https://doi.org/10.1016/j.pharmthera.2015.04.002
Wassmann K, Benezra R (2001) Mitotic checkpoints: from yeast to cancer. Curr Opin Genet Dev 11(1):83–90. https://doi.org/10.1016/s0959-437x(00)00161-1
Wu ZH, Yang DL (2020) High CENPM mRNA expression and its prognostic significance in hepatocellular carcinoma: a study based on data mining. Cancer Cell Int 20:406. https://doi.org/10.1186/s12935-020-01499-y
Wu Y, Ginther C, Kim J, Mosher N, Chung S, Slamon D et al (2012) Expression of Wnt3 activates Wnt/β-catenin pathway and promotes EMT-like phenotype in trastuzumab-resistant HER2-overexpressing breast cancer cells. Mol Cancer Res 10(12):1597–1606. https://doi.org/10.1158/1541-7786.Mcr-12-0155-t
Xiao Y, Najeeb RM, Ma D, Yang K, Zhong Q, Liu Q (2019) Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res 38(1):458. https://doi.org/10.1186/s13046-019-1444-0
Yang P, Pei X, Deng J, Li X (2022) Comprehensive analysis of centromere protein family member genes in lung adenocarcinoma. Crit Rev Eukaryot Gene Expr 32(4):57–72. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2022040641
Yu H (2002) Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol 14(6):706–714. https://doi.org/10.1016/s0955-0674(02)00382-4
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M et al (2022) Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer 21(1):144. https://doi.org/10.1186/s12943-022-01616-7
Zheng C, Zhang T, Li D, Huang C, Tang H, Ni XF et al (2020) Upregulation of CENPM facilitates tumor metastasis via the mTOR/p70S6K signaling pathway in pancreatic cancer. Oncol Rep 44(3):1003–1012. https://doi.org/10.3892/or.2020.7673
Zhou H, Bian T, Qian L, Zhao C, Zhang W, Zheng M et al (2021) Prognostic model of lung adenocarcinoma constructed by the CENPA complex genes is closely related to immune infiltration. Pathol Res Pract 228:153680. https://doi.org/10.1016/j.prp.2021.153680
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Q.K., L.W., and J.L. were collectively responsible for the comprehensive research design. J.L. and L.W. performed the experiments and wrote the manuscript. H.W. and X.C. assisted with the bioinformatics analysis and data visualization. Q.K. reviewed the article and made revisions. All authors contributed to the article and approved the final submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, L., Li, J., Wang, H. et al. Identification of the oncogenic role of centromere protein M in non-small cell lung cancer via CDC20/MYBL2/Wnt signaling pathways. J Mol Histol 56, 144 (2025). https://doi.org/10.1007/s10735-025-10423-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10735-025-10423-5