Abstract
Introduction
TRNA-derived small RNAs(tsRNAs) play an important role in many biological processes, and their dysregulation is closely related to the progression of cancer, but the research trend and future direction are not clear. This study aims to identify the leading contributors, collaboration networks, and emerging research trends in tsRNAs and their role in oncology, providing a more comprehensive and intuitive reference for researchers in this field.
Materials and methods
Related publications related to tsRNA in the field of oncology from 1990 to 2022 were collected from the Science Citation Index Expanded through the Web of Science Core Collection (WOSCC) database on 6 December 2022.
Results
There were 2,108 publications related to tsRNAs in oncology. The articles came from 69 countries/regions, 2,218 institutions, 11,340 authors, and 200 journals, and included 9,530 keywords. The annual total number of papers and total global citation score increased steadily every year over the study period. Among the articles related to tsRNAs in oncology, the United States had the highest number of publications with 732 articles, and the United States, China, Japan, Canada, and South Korea had the highest number of collaborations. Seoul National University Sun and the journal Nucleic Acids Research had the most publications at 81 and 63 articles, respectively, and the keyword “tRF” was a hotspot.
Conclusion
This study provides an in-depth analysis of the research status and development trends of tsRNAs in the field of cancer from a bibliometric perspective. Offering possible guidance for researchers to explore hot topics and frontiers, select suitable journals, and partners in this field.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Avoid common mistakes on your manuscript.
1 Introduction
Cancer is one of the leading causes of human health [1].There are an estimated 19.3 million new cancer cases and nearly 10 million cancer deaths worldwide, and a projected global cancer burden of 28.4 million cases in 2040, a 47% increase from 2020, according to Global Cancer Statistics 2020 [2].
Transfer RNAs (tRNAs) are traditionally considered bridging molecules that help the ribosome decode messenger RNAs (mRNAs) and synthesise proteins. Recent studies have shown that tRNA-derived small RNAs (tsRNAs) are a new class of non-coding sRNAs derived from mature or precursor tRNA, which divided into two categories: tRNA-derived stress-induced RNAs (tiRNAs) and tRNA fragments (tRFs). The relative abundance of tsRNAs is depending on the expression of tRNA fragments in tissue, and disease [3]. The expression of tsRNAs varies widely among different biological fluids [4] and is more stable and not easily degraded [5]. They have miRNA-like functions in regulating gene expression at the post-translation and immune response, cell activity, and other biological functions.
As functional regulatory molecules, tsRNAs are aberrantly expressed in a variety of tumours, such as gastric, lung, breast, colorectal, liver, and prostate cancers, and can be used as potential biomarkers and therapeutic targets [4].
At present, there are some studies on tsRNAs in the field of cancer, but there is a lack of research on predicting the future technological innovation and direction of development in this field. Therefore, with the help of CiteSpace and VOSviewer visualisation tools, This study aims to identify the leading contributors, collaboration networks, and emerging research trends in tsRNAs in oncology.
2 Materials and methods
2.1 Data source and search strategy
Related publications from 1 January 1990 to 6 December 2022 were collected from the Science Citation Index Expanded through the Web of Science Core Collection (WOSCC) database on 6 December 2022. With the search topics “cancer” OR “tumour” OR “neoplasm” AND “tRNA” OR “tsRNA” OR “tiRNA” OR “tRF” and the literature types “article” and “review”, a total of 2,108 articles satisfying these search criteria were identified for further analysis.
2.2 Analytical methods
Data analysis and data visualisation were performed using CiteSpace and VOSviewer version 1.6.14. CiteSpace is a software developed by Prof. Chaomei Chen and applied to the visual analysis of scientific literature, which can analyse changes in literature and the frontier of discipline hotspots [6, 7]. VOSviewer software, developed by the Science and Technology Research Center of Leiden University, focuses on bibliometric networks, such as co-citation, co-authorship, and keyword co-occurrence. Overall, these analytical tools provided an objective and diverse perspective on tsRNAs research in the field of oncology.
3 Results
3.1 General statistics
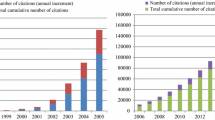
From 1 January 1990 to 6 December 2022, 2,108 tumour-related publications that discussed tsRNAs were available in the WOSCC database. We found articles from 69 countries/regions, 2,218 institutions, 11,340 authors, 200 journals, and 9,530 keywords. There were two types of publications: 1,828 research articles (86.72%) and 280 review articles (13.28%). As shown in Fig. 1, from 1990 to 2022, the cumulative number of published papers surged from 3 to 237, while total global citation score from 0 to 8,459 over the same period. A pivotal turning point occurred in 2015, when both metrics entered a phase of exponential growth, with annual publication output and citation accumulation increasing at unprecedented rates thereafter.
3.2 Country/Region characteristics
A total of 69 national/regional research groups have published articles related to tsRNAs in tumours during the period of 1990 to 2022.Overall, the countries/regions with the largest number of global publications are mainly concentrated in North America, East Asia and some parts of Europe (Fig. 2A). During the research period, the United States published the most articles on tsRNAs in tumors, with 732 articles. The People's Republic of China, Japan, South Korea and the United Kingdom (UK) followed (Fig. 2B). From the perspective of the growth trend, the number of publications in the United States has gradually and steadily increased; the number of publications in China has grown rapidly since 2012, while the number of publications in Japan has remained stable (Fig. 2C).
Major countries/regions where tsRNAs are distributed in oncology publications A Geographic distribution of tsRNAs in oncology. B The top 10 countries in tsRNA publication are distributed among oncology publications and their corresponding global total citation scores. C Annual trends for the top 3 most productive countries in the publication
3.3 Academic collaboration
Academic collaborations between different countries/regions, institutions, and authors often facilitate the exchange of knowledge, thus broadening the horizons of the field. As expected, in the field of tsRNAs in oncology, important partnerships occur at multiple levels (Fig. 3). Our study performed visualisation of the cooperative network using VOSviewer. Cluster analysis is a data mining technology that determines the topic distribution and organisation structure in the knowledge domain by obtaining multiple structured clusters [8]. 69 countries/regions collaborated. The two largest nodes are the Unites States and China, and the United States, China, Japan, Canada, and South Korea had the largest number of collaborations. Additionally, there is a high level of cooperation among other countries (Fig. 3A).
Academic collaboration on tsRNAs in the oncology field publications. A Academic collaboration between different countries/regions. B Collaboration between different institutions. C Collaboration between different authors. Each node represents a different country/region, institution, or author, the size of the node represents the number of publications, the distance between nodes represents the closeness of cooperation, and the thickness of the connections between nodes represents the amount of cooperation. Each colour represents a cluster, i.e., a group of projects in a network with similar properties
Another useful method of assessing the status of collaboration is to examine partnerships between organisations. Out of the 2,218 organisations, 168 met the threshold of at least six documented collaborations.The organizational collaboration network revealed four dominant clusters: a purple cluster centered at Seoul National University Sun, a blue cluster led by Harvard University, a green cluster anchored by the U.S. National Cancer Institute, and a red cluster spearheaded by Sun Yat-sen University. While intra-cluster interactions were frequent and dense (evident within each color-coded group), cross-cluster exchanges were sparse, suggesting a fragmented yet coordinated multi-institutional collaboration pattern with room for enhanced inter-cluster integration (Fig. 3B). In terms of organizational collaboration, Seoul National University Sun, National Institutes of Health, and the University of California system occupy the top three positions (Table 1).
To identify the most influential experts in oncology research involving tsRNAs over the past 32 years, authors were ranked based on their publication count and total link strength. Among the 11,340 researchers analyzed, 43 individuals exhibited frequent collaborative activity (> 9 collaborations), with Kim Sunghoon being the most prominent contributor and central hub in the collaboration network. Notably, Kwon Yoon Soo, Han Jung Min, and Yoon Ina also made substantial contributions and played pivotal roles in the collaborative among researchers (Fig. 3C).
3.4 Journal performance
In total, 2,108 papers were published in 200 journals. The top 10 journals with the highest number of publications had 332 papers (Table 2), accounting for 15.7% of all articles. Five journals are based in the United States, two in the UK, one in Greece, one in the Netherlands, and one in China. The impact factor (IF) is a presumed marker of journal impact, based on the frequency with which journal articles are cited by other scientific publications. The top three journals with the highest IF in 2021 were Nucleic Acids Research, Cancer Research, and Proceedings of the National Academy of Sciences of the United States of America.
3.5 Keyword detection and burst analysis
Keywords can reflect the frontiers and hotspots of a certain field. Among the 9,530 keywords, 110 were selected for cluster analysis with a minimum of 25 co-occurrence times, and a total of five clusters emerged. VOSviewer was used to cluster keywords for visualisation. In Fig. 4, the five clusters are indicated by different colours: (1) red for represents the effect of tsRNA gene expression on disease studies (“gene expression”, “growth”, “apoptosis”); (2) green for tsRNA source studies (“tRNA”, “reveals”, “stress” and “identification”); (3) blue for mechanism studies (“cancer”, “mechanisms” and “gene”); (4) yellow represents experimental studies of tsRNA phenotype (“proliferation”, “migration”, “progression” and “invasion”); (5) purple represents tsRNA-mediated protein expression studies (“protein”,“mutation” and “transfer-RNA”).
In order to analyse the research hotspots and future development trends of tsRNAs in the field of oncology, CiteSpace was used to detect the burst of keywords based on the frequency of keywords and citations, and analyse the changes in keywords over time. The results showed that since 1991, the keywords “carcinoma”, “nucleotide sequence”, and “DNA” have gradually evolved. As of 2020, the keywords with consistently high interest are “tRNA-derived fragment” (2018–2022), “small non-coding RNA” (2019–2022), “promoting cell proliferation” (2020–2022), and “gastric cancer” (2020–2022) (Table 3).
3.6 Co-citation analysis
Paper citation frequency is an important index for measuring the social influence in bibliometrics. Table 4 lists the top five most-cited papers in the WOS database. As evident from the large number of cited studies, the regulation of tsRNAs in tumour gene expression is currently of great interest to researchers. In addition, it is still an important research topic to explore tRF as a potential novel and stable biomarker for tumour diagnosis and its influencing factors.
The clustering of co-cited papers was divided into six time periods, and this clustering can indirectly reveal research hotspots in the field at different time periods (Fig. 5). The circles in the figure are termed tree ring histories, and the size of a ring reflects the number of times the paper has been cited. The colour of each circle represents the corresponding year; the closer it is red, the closer the year is to the present. In recent years, tsRNAs have been studied more frequently (red circles) in “#0 tRNA-derived fragments”.
4 Discussion
4.1 Main research hotspots
We performed a bibliometric analysis of 2,108 studies on tsRNAs in oncology. Papers published from 1990 to 2022 were retrieved from the WOSCC database using multiple literature analysis software and computational algorithms. This paper summarises tsRNA research leading contributors, collaboration networks, and emerging research trends in tsRNAs and their role in oncology.
In 2019, the WHO reported that cancer incidence and mortality are increasing rapidly worldwide [1, 2], therefore, in-depth research on cancer screening, early diagnosis, molecular mechanisms, targeted therapy, and other related fields is urgently needed to improve the cancer diagnosis rate, improve quality of life, and delay mortality. Additionally, tRNA is generally regarded as a steward molecule, which is involved in protein translation mainly by transporting amino acids to ribosomes and plays an important role in the occurrence. With the development of high-throughput sequencing technology, increasing evidence has shown that tRNA and its derived fragments are involved in a variety of molecular processes in tumours, including stress particle formation,proliferation, apoptosis, mRNA stabilisation,and translation regulation [9].
In terms of the countries where we have tsRNA published in cancer research,the United States, China, and Japan ranked among the top three. Regarding the United States, tRNA has been studied previously. In 1965, Holley Robert William isolated tRNA for the first time and clarified its sequence and general structure [10]. In 1982, Professor Yuet Wai Kan proposed the treatment of β-thalassaemia by blocking tRNA [11]. Recent studies have shown that changes in tRNA abundance or function alter tRNA activity and reprogram mRNA translation, leading to the occurrence of human tumours. The United States has rigorous and sensitive scientific research teams, with good experimental conditions and equipment. As the leader of tsRNA research in the field of cancer, this has led to considerable development of this research. Compared to the United States, China has a relatively low incidence of cancer, but the overall population base is large, the overall incidence is high, and the cancer mortality and disability adjusted life year annual rates are higher than those of the United States [12]. In recent years, with the development of China's economy and society, the Chinese government has increased its economic investment in medical research and has promoted a group of research teams dedicated to the field of cancer. As a result, tsRNA-related publications in the field of cancer and the corresponding total global citation score have increased significantly in the past decade. Japan has conducted earlier research in the field of cancer, particularly gastric cancer. Since 1960, early cancer screening, standardised surgical lymph node dissection, and preoperative neoadjuvant and postoperative chemotherapy have improved the 5-year survival rate of patients with gastric cancer to 52%, higher than the 25% rate in Western countries, such as the United States [13, 14].
In terms of scientific collaboration, the top three countries with the highest number of published papers (the United States, China, and Japan) had the highest number of close collaborations, as seen in the VOSviewer visual collaboration network graph. Among them, the distances between the Unites States and China, Japan, South Korea, and Israel are approximately the same, and the lines between the United States and China, South Korea, Japan, and the UK are thicker, indicating that the United States has good academic cooperation and close relationships with these countries. The distances between China and South Korea, Japan and Iran, and the UK and Germany indicate close cooperation.
The number of citations and the H-index reflect the influence of an organisation. Among the top 10 publications, six were from the United States, two from France, one from South Korea, and one from China; the top three institutions were Seoul National University Sun, National Institutes of Health, and the University of California system. As a result of the increase in the total number of articles published in the United States, the number of articles published by American institutions also increased. Additionally, four clusters led by Seoul National University Sun, Sun Yat Sen University, Harvard University, and the U.S. National Cancer Institute were formed, having close communication and cooperation with each other. A total of 43 high-producing authors with more than eight publications were divided into three groups, and the closely intertwined intersections between these groups indicated a tight network of collaboration among scientists worldwide. Kim Sunghoon (Total link strengtr = 103) of Yonsei University in South Korea has made outstanding contributions to tsRNA research and author collaboration.
Literature co-citation means that two articles appear in a third’s cited bibliography simultaneously and reflects the close relationship between published articles. Based on this, further co-citation clustering analysis can be performed. American scholar David H. Munn reported that uncharged tRNA elevated the activation of the general control nonderepressible 2 (GCN2) pathway in T cells and that T cells targeting the disruption of GCN2 were insensitive to indoleamine 2,3-dioxygenase (IDO) on plasmacytoid-like dendritic cells. In contrast, immunoregulatory dendritic cells expressing IDO are associated with tumour, autoimmune, and transplantation tolerance [15]. Tisdale MJ reported that the activation of dsRNA-dependent protein kinase was involved in decreased protein synthesis cancer cachexia. This activation reduces methionyl-tRNA binding to the 40S ribosomal subunit by increasing phosphorylation of eIF2α.Consequently, it enhances expression of the ubiquitin–proteasome pathway through activation NF-ĸB [16]. Yong Sun Lee identified a group of tRNAs-derived small RNAs using ultra-high-throughput sequencing, which ranked second in abundance. Among these tRFs, a particular variant known as tRF-1001 exhibited high expression levels in prostate cancer cell lines. It derived from the precursor transcript of Ser-TGA tRNA and its expression was association with cellular proliferation [17]. Hong and colleagues found that glucocorticoid receptor interacting protein 1 (GRIP1) interacts with the hormone-binding domain (HBD) of the glucocorticoid receptorand overexpression of GRIP1fragment in mammalian cells did not disrupts the expression of the tRNA gene promoter [18]. From the co-cited literature, it is clear that tsRNAs have become a hotspot in oncology research in recent years, and enhancing tRNA knowledge has broad development and application in oncology. Furthermore, the purpose of citation analysis is to determine the impact of a study by counting the number of times it is cited. The current findings show that the topic of tsRNAs has the highest citation frequency in basic oncology research, and that Nucleic Acids Research has the frequently cited journal within oncology.
4.2 Future research trends
The clustering of co-cited literature indirectly shows research hotspots in this field at different time periods. Keywords are the core summary of an article, and high-frequency keywords can reflect research hotspots and trends [19]. Keyword analysis can better reflect the research hotspots of tsRNAs in tumours. From 1991 to 2022, keywords like “carcinoma”, “nucleotide sequence”, and “DNA”, gradually evolved into "tRNA-derived fragments”, “small non-coding RNA”, and “promoting cell proliferation”. The evolution process from the keywords is tsRNA understanding development process, early only considered tsRNA as a tumor participant, while recently it is believed that its tsRNA cleavage products regulate tumorigenesis and development, indicating that the studies of its mechanism is the future of tsRNA development.
“Aminoacyl tRNA synthetase” has appeared as a clustering keyword over several years, indicating that aminoacyl tRNA synthetase has been a key research focus for scholars from various countries, while in recent years, more attention has been paid to the research of tRF in tumours. Most tRFs are generated by the cleavage of angiogenin (ANG). The expression of ANG is increased in almost all cancers, which further promotes the increase of tRF expression [20]. tRF mainly inhibits protein translation by preventing peptide bond formation [5] or acting as a signal transducer [21] or binds to RNA-binding proteins [22] to regulate mRNA stability, modulate reverse transcription and regulate apoptosis, thereby controlling the cell processes and the occurrence and development of cancer [23].
4.3 Limitations
This study provides valuable insights into the hotspots and developmental trends of tumor-related tsRNA research through bibliometric analysis. However, several limitations warrant acknowledgment. First, the study relies primarily on data from the WOSCC database. While WOSCC is a well-established source, its exclusion of other databases and non-English publications may compromise the global representativeness of the findings. For instance, emerging studies from Asia–Pacific regions or specialized journals focused on RNA biology might remain underrepresented, Literature of real value may be missed. Second, bibliometric analyses inherently depend on quantitative metrics such as publication counts and citation numbers. This limitation is particularly salient in rapidly evolving fields like tsRNA research, where recently published high-impact studies may not yet have accumulated sufficient citations to reflect their true significance. Consequently, the conclusions drawn from this study should be viewed with caution, taking these limitations into account. Further empirical studies and validation eforts will be essential to strengthen the fndings and to extend the insights provided by this analysis into practical clinical frameworks.
Moreover, bibliometric analysis mainly relies on specific keywords, which may affect the accuracy and completeness of the results. Additionally, although tsRNA shows considerable promise in tumors, it is still in the early stage of theoretical research. Its potential for clinical application and transformation needs further validation, and the path of conventional clinical application may be challenged by the complexity of the real world.
5 Conclusion
This study provides an in-depth analysis of the research status and development trends of tsRNAs in the field of cancer from a bibliometric perspective. Offering possible guidance for researchers to explore hot topics and frontiers tsRNAs in cancer, select suitable journals, and partners in this field.
Data availability
The original contributions to this study are included in the article material. Further enquiries can be directed to the corresponding author.
Abbreviations
- tRNA:
-
Transfer RNA
- tsRNAs:
-
TRNA-derived small RNAs
- WOSCC:
-
Web of Science Core Collection
- WHO:
-
World Health Organization
- mRNAs:
-
Messenger RNAs
- tRFs:
-
TRNA fragments
- tiRNAs:
-
TRNA-derived stress-inducible RNAs
- IF:
-
Impact factor
- GCN2:
-
General control nonderepressible 2
- IDO:
-
Indoleamine 2,3-dioxygenase
- mtDNA:
-
Mitochondrial DNA
- HBD:
-
Hormone-binding domain
- GRIP1:
-
Glucocorticoid receptor interacting protein 1
- UK:
-
The United Kingdom of Great Britain and Northern Ireland
- ANG:
-
Angiogenin
References
World Health Organization (WHO). Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2019. WHO.2020. who.int/data/gho/data/themes/morta lity-andglobal-health-estimates/ghe-leading causes-of-death. Accessed 11 Dec 2020.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Shao Y, Sun Q, Liu X, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90(5):730–8. https://doi.org/10.1111/cbdd.12994.
Balatti V, Pekarsky Y, Croce CM. Role of the t RNA- derived small RNAs in cancer: new potential biomarkers and target for therapy. Adv Cancer Res. 2017;135:173–87. https://doi.org/10.1016/bs.acr.2017.06.007.
Sobala A, Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553–63. https://doi.org/10.4161/rna.24285.
Chen C, Morris S. Visualizing evolving networks: minimum spanning trees versus pathfinder networks. Seattle: IEEE; 2003. p. 67–74.
Chen C. Citespace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci. 2006;57(3):359–77. https://doi.org/10.1002/asi.20317.
Lin H, Zhu Y, Ahmad N, Han Q. A scientometric analysis and visualization of global research on brownfields. Environ Sci Pollut Res. 2019;26(17):17666–84. https://doi.org/10.1007/s11356-019-05149-3.
Tosar JP, Cayota A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020;17(8):1149–67. https://doi.org/10.1080/15476286.2020.1729584.
Temple GF, Dozy AM, Roy KL, Kan YW. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature. 1982;296(5857):537–40. https://doi.org/10.1038/296537a0.
Buvoli M, Buvoli A, Leinwand LA. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol Cell Biol. 2000;20(9):3116–24. https://doi.org/10.1128/MCB.20.9.3116-3124.2000.
Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun. 2021;41(10):1037–48. https://doi.org/10.1002/cac2.12197.
Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I; EUROCARE-4 Working Group. Recent cancer survival in Europe: a 2000–02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007 Sep; 8(9):784–96. https://doi.org/10.1016/S1470-2045(07)70246-2. Erratum in: Lancet Oncol. 2008 May; 9(5):416. PMID: 17714993.
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108.
Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, Mellor AL. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22(5):633–42. https://doi.org/10.1016/j.immuni.2005.03.013.
Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89(2):381–410. https://doi.org/10.1152/physrev.00016.2008.
Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639–49. https://doi.org/10.1101/gad.1837609.
Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93(10):4948–52. https://doi.org/10.1073/pnas.93.10.4948.
Liu XM, Zhao J, Xu JM. Research on immobilization of heavy metals in contaminated agricultural soils—bibliometric analysis based on Web of Science database. Acta Pedol Sin. 2021;58(2):445–55.
Yu X, Xie Y, Zhang S, Song X, Xiao B, Yan Z. tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics. 2021;11(1):461–9. https://doi.org/10.7150/thno.51963.
Czech A, Fedyunin I, Zhang G, Ignatova Z. Silent mutations in sight: co-variations in tRNA abundance as a key to unravel consequences of silent mutations. Mol Biosyst. 2010;6(10):1767–72. https://doi.org/10.1039/c004796c.
Zhu L, Ge J, Li T, Shen Y, Guo J. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–7. https://doi.org/10.1016/j.canlet.2019.03.012.
Gupta T, Malkin MG, Huang S. tRNA function and dysregulation in cancer. Front Cell Dev Biol. 2022;10:886642. https://doi.org/10.3389/fcell.2022.886642.
Acknowledgements
The authors would like to thank Dr. Mingtao Zhang, the Second Hospital of Lanzhou University, and Lanzhou University Library for supporting the work.
Funding
Science and Technology Planning Project of Gansu Province (21CX6FD163 and 25JRRA255); Science and Technology Planning Project of Baiyin City(2024-1-20Y), The Second Group of Longyuan Young Talents in Gansu Province (2023-11).
Author information
Authors and Affiliations
Contributions
This work was conceived by Z-HG. Data was collected and downloaded by J-HW. The visualization work was performed by E-LS. The manuscript was written by J-HW and Z-HG revise the manuscript and proposed constructive opinions. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, J., Shao, El. & Gao, Z. Emerging trends and hotspots of tRNA-derived small RNAs in tumours: a bibliometric analysis via VOSviewer and CiteSpace. Discov Onc 16, 767 (2025). https://doi.org/10.1007/s12672-025-02628-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-025-02628-7