Abstract
Cytoplasmic mRNA decay is effected by exonucleolytic degradation in either the 5’ to 3’ or 3’ to 5’ direction. Pervasive terminal uridylation is implicated in mRNA degradation, however, its functional relevance for bulk mRNA turnover remains poorly understood. In this study, we employ genome-wide 3’-RACE (gw3’-RACE) in the model system fission yeast to elucidate the role of uridylation in mRNA turnover. We observe widespread uridylation of shortened poly(A) tails, promoting efficient 5’ to 3’ mRNA decay and ensuring timely and controlled mRNA degradation. Inhibition of this uridylation process leads to excessive deadenylation and enhanced 3’ to 5’ mRNA decay accompanied by oligouridylation. Strikingly we found that uridylation of poly(A) tails and oligouridylation of non-polyadenylated substrates are catalysed by different terminal uridyltransferases Cid1 and Cid16 respectively. Our study sheds new light on the intricate regulatory mechanisms underlying bulk mRNA turnover, demonstrating the role of uridylation in modulating mRNA decay pathways.
Similar content being viewed by others
Introduction
Messenger RNA turnover is a fundamental process in eukaryotic cells that ensures the dynamic regulation of gene expression. The majority of eukaryotic mRNAs produced in the nucleus are constantly degraded in the cytoplasm. This transient nature of mRNA molecules is necessary for gene expression flexibility1. All mRNAs in the cytoplasm are believed to be degraded by the same conserved pathways2. The most important determinants of mRNA stability are specific terminal features, namely the 5’ cap and 3’ polyadenosine (poly[A]) tail. These structural elements play important roles in mRNA protection, translation initiation, and decay. Bulk mRNA decay is usually initiated by the poly(A) tail shortening, catalysed by specific deadenylase complexes3. Following deadenylation, mRNA degradation can occur from either the 5’ end or the 3’ end, mediated by distinct exonucleases.
Both 3’ and 5’ directed decay pathways have been thoroughly studied in budding yeast. During 5’ to 3’ decay, the 3’-end of the deadenylated transcript is bound by the Lsm 1-7 complex, which activates the removal of the protective 5’-cap structure and leaves the mRNA molecule accessible for cytoplasmic 5’ to 3’ exonuclease, Xrn12. 3’ to 5’ decay is executed by the multi-subunit exosome complex and facilitated by the auxiliary SKI complex4. Both exonucleolytic decay pathways serve redundant functions and can complement each other5. There is evidence that the 5’ to 3’ decay pathway is dominant6. However, the exact contributions to, and determinants of decay directionality are not well established.
Recent research has shed light on the role of polyadenosine tail uridylation, a conserved modification of mRNA, in mRNA decay. Uridylation involves the addition of one to a few uridine residues to the end of the poly(A) tail, and it has been shown to influence mRNA stability and decay rates7,8. Poly(A) tail uridylation is a conserved feature of the eukaryotic transcriptome, with the notable exception of budding yeast9. Uridylation is catalysed by a specific class of non-canonical polymerases - terminal uridyltransferases (TUT-ases)9,10. Despite the conservation of uridylation across eukaryotes, the exact function of mRNA uridylation in general mRNA turnover remains poorly understood. Several consequences of uridylation for mRNA fate have been described. Uridylation was shown to destabilise mRNA by facilitating Lsm1–7 complex binding and thus promoting degradation through the 5’ to 3’ pathway7,8. In plants, uridylation additionally protects mRNA from excessive deadenylation and inhibits the activity of CAF1 deadenylase in vitro11,12. Uridylation can also trigger 3’ to 5’ mRNA decay by conserved uridylation-dependent exonuclease Dis3l213. Strikingly, eliminating uridylation in yeast does not result in strong phenotypes in standard growth conditions. Moreover, uridylation is difficult to study on a genome-wide level, with only a handful of methods dedicated to its high-throughput detection14.
In this study, we aimed to unravel the role of mRNA uridylation in bulk mRNA decay using fission yeast (Schizosaccharomyces pombe) - a model organism with well-established mRNA uridylation8,15. There are two annotated terminal uridyltransferase genes encoded in the fission yeast genome: cid1 and cid1616. Cid1 was shown to be responsible for the majority of poly(A) tail uridylation8, while Cid16 was proposed to be involved in both mRNA uridylation17 and the turnover of Argonaute-bound small RNAs18.
To obtain a comprehensive understanding of the uridylation landscape of fission yeast we applied genome-wide 3’RACE, a novel technique for mapping RNA 3’-end architecture. By using a collection of mRNA decay mutants and investigating the contributions of Cid1 and Cid16 to uridylation in different genetic backgrounds, we gained insights into the roles of these enzymes in directing mRNA decay towards specific pathways. In particular, we discovered that two fission yeast TUT-ases catalyse two distinct types of uridylation: mRNAs with shortened poly(A) tails are uridylated by Cid1, while completely deadenylated mRNAs are subjected to oligouridylation by Cid16. Cid1-mediated uridylation routes decay towards the 5’ to 3’ pathway, while Cid16-mediated oligouridylation facilitates 3’ to 5’ degradation. Elimination of cid1 boosts 3’ to 5’ directed decay and is accompanied by the accumulation of deadenylated mRNAs that become targets for Cid16 mediated oligouridylation.
Results
Genome-wide 3’-RACE describes architecture of RNA 3’-ends
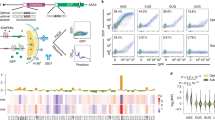
To gain a comprehensive picture of 3’-end transcript architecture we established a genome-wide 3’-RACE method (gw3’-RACE) (Fig. 1a). In short, an adaptor was ligated to the 3’-end of rRNA-depleted total RNA and an approach similar to very efficient Bulk RNA Barcoding and sequencing strategy (BRB-seq)19 was used to produce sequencing libraries. Libraries were paired-end sequenced using the Illumina NOVA-seq platform with read 1 (R1) aligning to the transcript body and read 2 (R2) covering the sequence of the 3’ extremity of this transcript, including the non-templated nucleotides. This strategy appeared to be robust, fast and cost-effective, allowing the production of good quality sequencing libraries covering transcripts 3’-end region (Fig. 1b and Supplementary Fig. 1a–c). It was recently demonstrated that Illumina sequencing platforms can accurately estimate poly(A) homopolymer size within the range of poly(A) tail lengths in yeast20. We used a series of spike-ins with poly(A) stretches from 10 to 40 nucleotides and confirmed that we could correctly estimate homopolymer length in our sequencing data (Fig. 1d).
a schematic overview of gw3’-RACE protocol. b an example of read coverage obtained from RNA-seq and gw3’-RACE experiment. c density plot quantifying the varying-length adenosine homopolymers that were spiked into NovaSeq libraries. Expected lengths are indicated with dashed lines. d the architecture of 3’-ends in different RNA classes. Depicted is a mean fraction of each type of 3’-end detected for genes with more than 20 mapped reads and calculated for each class of RNA (average of two repeats). n equals the number of genes passing the read count threshold. e correlation of mean tail length per protein coding genes obtained for two biological repeats. Depicted are genes with more than 20 tailed reads. f correlation of mean tail length per protein-coding gene detected in our data (average of two repeats) with mean tail length detected using the PAL-seq method for fission yeast mRNAs21. g distribution of 3’-end positions of different 3’-end types relative to transcription end site (TES).
We next applied custom data analysis to retrieve information about the status and mapping position of the 3’-ends (Supplementary Figs. 1e, 2). Our results align with existing knowledge regarding the 3’-end architecture of various RNA classes, showing that most structural RNAs lack tails while coding transcripts exhibit extensive tailing (Fig. 1d). Over 95% of all R2 reads fell into one of the four RNA 3’-end types: (i) poly(A) tails, (ii) uridylated poly(A) tails (iii) pure oligouridine tails or (iv) without non-templated additions referenced to as “no tails”, tails not matching any of the above classes are references as “others” (Fig. 1d). As expected poly(A) and uridylated poly(A) tails were found predominantly in mRNA and ncRNAs. A low level of oligouridylation was also detected for those species. Oligouridylation was, however, a prominent 3’-end modification of mitochondrially encoded RNAs (mtRNA) (Fig. 1d).
We found a good agreement between the mean mRNA tail length calculated for two biological repeats (Pearson’s correlation coefficient r = 0.73 for genes with at least 20 tailed reads) (Fig. 1e). The average tail length per gene also showed a significant positive correlation with tail lengths calculated using an alternative method (PAL-seq, Fig. 1f)21.
The 3’-end of all three kinds of tailed read [poly(A), poly(A)U and oligo(U)] mapped in the vicinity of the annotated mRNA transcription end sites (TES), while non-tailed reads mapped across the mRNA body (Fig. 1g).
Metabolic genes are extensively uridylated
The average number of uridylated poly(A) tails detected among all mRNA mapped-tailed reads was about 20% (Fig. 1d) which is higher than uridylation reported in humans22 or plants11. However, similar to other systems, uridylation targets predominantly mRNAs with short poly(A) tails (Fig. 2a, b). Tails shorter than 17 nucleotides were uridylated with a frequency approaching 50%, uridylation frequency dropped for tails between 17 and 40nt, reaching a minimal level for tails longer than 40nt (Fig. 2b). This step-wise change in frequency of poly(A) tails uridylation correlates with the size of poly(A) stretch required for binding Pabp protein (20–25nt), and is in agreement with proposed antagonistic role of Pabp in uridylation23.
a Distribution of lengths of tails mapped to mRNAs (average of two repeats). The amount of tails (y-axis) of a given length (x-axis) is indicated as a percentage in all tailed reads. A stacked bar plot was used to visualise the split of tails of a given length into three categories. The tail length equals the sum of all non-templated nucleotides. Depicted are tails of length of 2nt or more. b the percentage of poly(A) uridylation calculated per tail length for reads mapped to mRNAs. Depicted is the mean of two repeats (data as in a) +/- SD as shaded area. c Barplot depicting the fraction of tails (y-axis) with indicated number of uridines added (x-axis); left panel: Us added to poly(A) tails, right panel: Us in oligo(U) tails. Bars depict mean values +/- SE of four biological replicates. d The mean number of uridines (y-axis) added to adenine tails of different lengths (x-axis). Depicted is the mean of two repeats (data as in a) +/- SD as shaded area. e from left: the relationship between mean mRNA tail length (this study) and mRNA copy per cell (data from24). Tail length is an average of two repeats calculated for all genes with at least 20 tailed reads (n = 2611 genes); the relationship between mRNA uridylation frequency and mRNA copy per cell. mRNA uridylation frequency was calculated by dividing the number of poly(A)U reads by all tailed reads mapped to the given gene (average of two repeats); the relationship between mean mRNA tail length and mRNA uridylation frequency; the relationship between mRNA uridylation frequency and mRNA half-life (data from47). f KEGG categories enriched in the 25% of genes with mRNAs with shortest tails or 25% of genes with most highly-uridylated mRNAs. Results were obtained using gProfiler48; Probability was calculated using Fisher’s one-tailed test and adjusted using Bonferroni correction. g examples of tail length and uridylation distribution for genes with high and low poly(A) tail uridylation frequency. A stacked bar plot was used to visualise the split of tails of a given length in three categories: poly(A), poly(A)U and oligo(U).
Similar to the situation in human cells, poly(A) tail uridylation consisted predominantly of one or two uridine residues (Fig. 2c)22. A Number of uridines added to pre-existing poly(A) tails was highest in the case of shortest tails, and with A number increase, showed a steadily decreasing trend (Fig. 2d). Oligouridylation was rare in mRNA and amounted to less than 0.1% of all mapped reads. Oligo(U) tails added to mRNAs were longer on average than U-stretches added to pre-existing mRNAs adenine tails (Fig. 2c).
According to our data total mRNA tail length anticorrelates with mRNA copy per cell (Fig. 2e)24, which is in agreement with previous observations by others25. We calculated transcript uridylation frequency, which is the percentage of poly(A) uridylated reads among all tailed reads detected for a given transcript. The transcript uridylation frequency differed between transcripts and spanned from none to over 60% of uridylated poly(A) tails detected (Supplementary Fig. 3a, b). Transcripts with shorter tails tended to have higher poly(A) tail uridylation frequency (Fig. 2e). Transcript-wise poly(A) tail uridylation frequency was positively correlated with mRNA copy per cell (Fig. 2e). Contradictory to what was reported for human cells22, we recorded a weak positive correlation between poly(A) tail uridylation frequency and mRNA half-life (Fig. 2e). We concluded that poly(A) tail uridylation in fission yeast is a feature of highly abundant, short-tailed genes.
While short poly(A) tails tend to be highly uridylated, high transcript uridylation frequency is not simply a consequence of transcript poly(A) tail length. Different functional mRNA categories were enriched among genes which transcripts showed the highest uridylation frequency and those with the shortest tails (Fig. 2f). While mRNAs of the ribosomal subunits and nuclear pore complex were among transcripts with the shortest tails, we did not find these categories among the 25% of genes with the highest poly(A) tail uridylation. Instead, the top uridylated mRNAs were enriched in amino acid and carbon metabolism genes. Of note, transcripts of ribosomal subunit genes were still uridylated above all genes average (Supplementary Fig. 3c). Highly uridylated transcripts had a peculiar tail profile characterised by an accumulation of short poly(A) uridylated reads, while genes with low uridylation displayed Gaussian-type tail length distribution (Fig. 2g).
Cid1 and Cid16 catalyse the uridylation of different RNA substrates
We next constructed strains with one or both fission yeast TUTase genes deleted and performed genome-wide 3’-RACE. Deletion of the cid1 gene resulted in a 90% decrease in the number of detected poly(A)U 3’-ends, proving that Cid1 is the main enzyme responsible for poly(A) tail uridylation8 (Fig. 3a, b). Single deletion of the cid16 gene had no significant impact on poly(A) tails uridylation, however, double deletion of cid1 and cid16 genes resulted in a 98% drop in the number of detected poly(A)U tails – markedly more than in the case of the single cid1 gene deletion (Fig. 3a). The level of residual uridylation observed in TUT-ase double deletion strain was comparable to one detected when we applied gw3’-RACE to analyse budding yeast transcripts were uridylation is absent (Supplementary Fig. 3d). Our results suggests that Cid16 can perform a fraction of poly(A) tail uridylation, while it may be a byproduct in the context of cid1 deletion. The observed redundancy in poly(A) uridylation between both TUTases is also supported by our earlier observations17.
a Change in the fraction of reads with poly(A) uridylated tails in TUT-ases deletion strains. Results are presented as a fraction of the given category of reads in all reads. Depicted is the mean value calculated based on two (mutants) or four (wt) independent repeats +/- SD. b Changes of mRNA tails architecture in TUT-ase mutants. For each mutant, a mean fraction of each type of 3’-end detected in mRNA aligned reads is depicted. Common genes with more than 20 mapped reads in all mutants were considered (average of two repeats) (n = 1828). c Change in the fraction of reads with oligouridylated tails (oligo(U)) in different deletion strains. Results are presented in (a). d Distribution of mean mRNA tail lengths per gene detected in different TUT-ase deletion strains (genes with more than 20-tailed reads detected in all tested samples are considered). e Distribution of tail-length of gw3’-RACE derived reads mapped to mRNA in different strains (average of two repeats). Amounts of tails (y-axis) of a given length (x-axis) are indicated as percentage in all tailed reads detected. A stacked bar plot was used to visualise the split of detected tails of a given length into three categories. The tail length equals the number of all non-templated nucleotides (like in Fig. 2a). f For each TUT-ase deletion strain, the change in the percentage of reads with a certain tail length over the wild-type strain was calculated. The average fold change for the two repeats for each tail length is depicted as a line with a standard deviation indicated as a shaded area. g Change in the distribution of 3’-end mapping position relative to the transcription end site (TES) of non-tailed reads detected in different yeast strains. A normalised density plot was used with a maximum value of the y-axis adjusted to 1 in each plot.
Strikingly, the cid16 gene deletion caused almost complete loss of oligouridylation of 3’-ends (Fig. 3c). This effect was slightly enhanced in the double deletion (Δcid1Δcid16) suggesting some level of redundancy also in the case of oligouridylation. The highest number of oligo(U) tailed reads were detected for mitochondrial transcripts (Supplementary Fig. 4a), therefore, oligouridylation of mtRNA seems to be a main function of Cid16. However, 3’-end oligouridylation also targeted non-tailed mRNAs with Cid16 responsible for both mtRNA and mRNA oligouridylation (Supplementary Fig. 4b). Both mtRNA and mRNA oligo(U) tailed reads mapped closely to annotated transcription end sites (TES) (Supplementary Fig. 4c).
While the number of uridines in oligo(U) tails is higher than the number of uridines added to poly(A) tails, in the cid1 deletion strain, residual poly(A) uridylated tails contained a slightly greater number of uridines, while in the cid16 deletion strain, residual oligo(U) tails were clearly shorter (Supplementary Fig. 4d, e).
Altogether, we conclude that the preferred substrates for Cid1-catalysed uridylation are shortened poly(A) tails, while Cid16 adds uridine preferentially to non-tailed RNAs. Accordingly, the primary substrate of Cid16 is mitochondrial transcripts but it is also capable of oligo-uridylating non-tailed mRNAs.
Deletion of one or both uridyltransferases had a minor effect on the global mean tail length of mRNAs (Fig. 3d, and Supplementary Fig. 5a). In accordance with previous experiments, we also did not observe reduced cell growth in standard conditions after the loss of uridylation (Supplementary Fig. 5b). Moreover, TUTase deletion resulted in very limited changes to mRNA steady-state levels (Supplementary Fig. 5c). We therefore looked for more subtle molecular consequences of uridyltransferase deletion.
Lack of uridylation leads to excessive deadenylation of mRNAs
We analysed the changes in global tail profiles that illustrate amounts of all the tailed reads mapped to mRNA in each TUTase deletion strain (Fig. 3e). While the global tail profile did not change in the Δcid16 mutant, we noticed an increase in the fraction of short-tailed reads in Δcid1 and double deletion strains (Fig. 3e). This effect was evident when we compared the change in the percentage of detected reads with a tail of a given length between each mutant and the wild type strain (Fig. 3f). The fraction of reads with tails shorter than about 10 nucleotides increased around two times in cid1 deletion strain and in Δcid1Δcid16 strain, while there was no change in the cid16 deletion strain. The increase in detected short-tailed reads was higher when we considered only reads from 10% of genes with the highest poly(A) tail uridylation in the wild-type strain, whilst the distribution of tail lengths for 10% of genes with low uridylation remained unchanged (Fig. 3f).
Genome-wide 3’-RACE records both tailed and non-tailed reads. The fraction of non-tailed reads detected in wild-type and uridyltransferase mutant samples did not change significantly (t test of mean change between biological repeats) (Fig. 3c). We observed, however, changes in the distribution of 3’-end mapping positions of non-tailed reads, especially in the Δcid1 and the Δcid1Δcid16 strain. The density profile of the non-tailed reads 3’-ends mapping positions in those strains indicates that more reads map adjacent to the annotated transcription end site (TES) as compared to the wild-type or single Δcid16 deletion (Fig. 3g). This trend was not observed in reads from 10% of transcripts with low uridylation frequency, while it is evident for 10% of transcripts with high uridylation frequency (Fig. 3g). Accumulation of non-tailed reads in cid1 deletion strain is further confirmed by increase in number of oligouridylated mRNAs in cid1 deletion (Fig. 3e and Supplementary Fig. 4b).
Uridylation protects 3’-end by stabilising short-tailed mRNAs
Uridylation of short poly(A) tails attracts binding of the Lsm 1–7 complex to the mRNA 3’-end, which in turn stimulates decapping followed by 5’ to 3’ decay8,13. In agreement with this, in Δlsm1 strain, we observed an accumulation of poly(A) tail uridylation (Fig. 4a), and significant global tail shortening (Fig. 4b). Deletion of cid1 in a Δlsm1 strain background caused loss of poly(A) uridylation, however, the poly(A)U and oligo(U) fractions in Δlsm1Δcid1 strain were higher than in the cid1 deletion, which we attribute to an increased contribution of Cid16 (Fig. 4a). The Δlsm1 strain exhibited a slow growth phenotype, which was further enhanced in synthetic media. This suggests partial auxotrophy (Fig. 4c, d and Supplementary Fig. 6a). Interestingly, we observed that the growth phenotype of the Δlsm1 strain was partially reversed by cid1 gene deletion (Fig. 4c, d).
a Changes in the composition of mRNA 3’-end architecture in different strains. Data for common genes with more than 10 reads in both repeats for each strain (n = 1335 genes). b Distribution of mean mRNA tail lengths per gene for different strains for common genes with more than 10 reads detected in each sample (n = 1335). c Growth curves of yeast grown in YES media, depicted is an average growth curve for at least four repeats with standard deviation indicated by a shaded area. d colony growth of different strains in complete (YES) or synthetic (EMM) media. Yeast strains were plated in rows in a series of 5-fold dilutions. e Distribution of tail-length of gw3’-RACE derived fragments mapped to mRNA in different strains (average of two repeats). Amounts of tails (y-axis) of a given length (x-axis) are indicated as percentages in all tailed reads detected. A stacked bar plot was used (like in Fig. 2a), and data as in (a). f Distribution of tail-length of fragments mapped to mRNA in different strains as in (e), (average of two repeats). g Change in the distribution of 3’-end mapping positions relative to the transcription end site (TES) of non-tailed reads. Normalised density plot was used with the maximum value of the y-axis adjusted to 1 in each plot; data as in (a). h Relationship between uridylation frequency and transcript fold change in Δlsm1 relative to the wild-type strain. i Change in mRNA uridylation frequency in Δlsm1 strain (y-axis) compared to the wild-type strain (x-axis). j Volcano plot depicting changes of mRNA expression level in Δlsm1 relative to the wild-type strain. Padj was obtained by the Wald test and corrected using the Benjamini and Hochberg method. k KEGG categories significantly enriched among genes up-regulated in the Δlsm1 strain. Enrichment probability was calculated using Fisher’s one-tailed test, and Bonferroni correction was used to obtain adjusted p-values. l Pearson correlation heatmap describing the relationship between proteome or transcriptome changes and uridylation frequency in Δlsm1 strain. The correlation value is indicated as the number and colour of the heatmap square.
Lsm1 gene deletion resulted in the accumulation of poly(A) uridylated tails, between 8 and 14 nucleotides long (Fig. 4e). Those phenotypes stem from decapping inhibition which hinders 5’ to 3’ decay. Similar accumulation is observed in the strains with deletion of genes acting in the 5’ to 3’ decay pathway downstream of the Lsm1–7 complex – exonuclease xrn1 and decapping activator edc1 (Fig. 4f). Accumulation of shortened uridylated tails was observed for both lowly and highly uridylated genes (Fig. 4e), indicating that all transcripts undergo uridylation and Lsm1–7 complex binding. The clear peak of short uridylated poly(A) tails disappeared when cid1 was deleted in the Δlsm1 strain background. Instead, we observed a buildup of very short (< 10nt) poly(A) uridylated tails and oligouridylated 3’-ends (Fig. 4e), as well as non-tailed 3’-ends located near annotated transcription end site (TES) (Fig. 4g). Taken together, this suggests an increased level of extensive deadenylation of mRNA in Δcid1Δlsm1 strain.
Interestingly, changes in transcript levels in the Δlsm1 strain correlate with the frequency of poly(A) tail uridylation in this strain (Fig. 4h), which suggests a stabilising role of mRNA uridylation in this background. Indeed mRNA half-lives calculated for 5 tested transcripts were consistently higher in Δlsm1 than in Δcid1Δlsm1 strain (Welch t test p value < 0.007) (Supplementary Fig. 6b, c). Uridylation was recently shown to inhibit deadenylation in vitro11. This could explain the stabilisation and build-up of uridylated mRNAs in Δlsm1 strain. Poly(A) tail uridylation frequency in the lsm1 deletion strain was highest for transcripts of genes already exhibiting high uridylation in the wild-type strain (Fig. 4i). Accordingly, functional categories enriched among transcripts up-regulated in the Δlsm1 strain (Fig. 4j) were similar to those for genes with high poly(A) tail uridylation in wild type cells (Fig. 4k) - amino acid and glucose metabolism. Changes in poly(A) uridylation frequency also correlated with changes in protein levels in the Δlsm1 strain (Fig. 4l).
Uridylation regulates contribution of 3’ to 5’ decay in mRNA turnover
In budding yeast, non-tailed mRNAs are targeted by the exosome with the help of the conserved SKI complex. In S. pombe, the 3’ to 5’ mRNA decay pathway may be additionally supported by Dis3l2 exonuclease which was shown to preferentially target oligouridylated RNAs13. Single deletions of dis3l2 or ski2 genes had no significant impact on 3’-tails architecture (Fig. 5a), suggesting the minimal contribution of each of the 3’ to 5’ decay pathways to bulk mRNA decay. Interestingly, we noticed a much stronger effect when 3’ to 5’ decay was impaired in the background of the cid1 gene deletion. In those cases, deletion of either of 3’ to 5’ decay pathways resulted in the accumulation of non-tailed and oligo-uridylated 3’-ends (Fig. 5a), this effect was more evident in the Δcid1Δski2 strain. Deletion of the ski2 gene in a Δcid1 strain background also resulted in mRNA mean tail shortening (Fig. 5b). The Δcid1Δski2, but not Δcid1Δdis32 strain grew significantly slower than the control or parental strains (Fig. 5b, c). Observed effects were further enhanced when the 3’ to 5’ decay pathways were impaired in the Δcid1Δlsm1 strain background. Deletion of dis32 or ski2 genes in Δcid1Δlsm1 strain background resulted in a massive increase in the amount of non-tailed and oligouridylated 3’-ends detected (Fig. 5a). We also observed global tail shortening and a slow growth phenotype in both Δcid1Δlsm1Δdis32 and Δcid1Δlsm1Δski2 strains (Fig. 5c). Of note, in all combinations ski2 deletion appeared to have a stronger effect than dis32 gene deletion (Fig. 5a–d and Supplementary Figs. 9, 10).
a Changes in the composition of mRNA 3’-end architecture in different strains. Data for genes with more than 10 reads in both repeats for each strain (n = 1196 genes). b Distribution of mean mRNA tail lengths per gene for different strains for common genes with more than 10 reads detected in each sample (c) Growth curves of different yeast strains. Yeast was grown in YES media, depicted is an average growth curve for at least four repeats with standard deviation indicated by a shaded area. d Distribution of tail-length of gw3’-RACE derived fragments mapped to mRNA in different strains (average of two repeats). Amounts of tails (y-axis) of a given length (x-axis) are indicated as percentages in all tailed reads detected. A stacked bar plot was used to visualise the split of detected tails of a given length into three categories. The tail length equals the number of all non-templated nucleotides (like in Fig. 2a),; data as in (a). e Change in the distribution of 3’-end mapping positions relative to the transcription end site (TES) of non-tailed reads detected in different yeast strains. Normalised density plot was used with the maximum value of the y-axis adjusted to 1 in each plot; data as in (a).
Analysis of global tail profiles unravelled a more detailed picture of the change in mRNA 3’-ends (Fig. 5d). Single deletion of dis32 or ski2 genes had no obvious effect on tail profile while deletion of dis32 gene in Δcid1 strain background led to an increase in the number of detected poly(A) tails smaller than 10nt and oligo(U) reads. Accumulation of short-tailed and oligo(U) reads was further enhanced in Δcid1Δski2 strain, and to an even greater extent in the Δcid1Δlsm1 strain. Accumulation of short-tailed and oligo(U) 3’-ends in the investigated mutants correlated with an increase in the number of non-tailed reads mapping in proximity to the transcription end site (TES) indicating accumulation of non-tailed mRNAs. This effect was again strongest for Δcid1Δski2 and Δcid1Δlsm1Δski2 strains (Fig. 5e).
Overall, we found that the contribution of 3’ to 5’ pathways to mRNA degradation in standard conditions is either minimal or very efficiently substituted by other pathways. However, when uridylation is compromised, the contribution of 3’ to 5’ decay to mRNA turnover grows. The increased number of detected oligo(U) 3’-ends in 3’ to 5’ decay pathway mutants suggests that oligouridylation assists in the 3’ to 5’ decay process. Both molecular and physiological phenotypes indicate that out of two 3’ to 5’ decay routes, exosome degradation is dominant, however, Dis3l2 can evidently contribute to the process.
Discussion
We developed a robust approach called genome-wide 3’-RACE to investigate the relationship between uridylation and different mRNA decay pathways. We demonstrated the utility of our approach for yeast material, where average tails are significantly shorter than in their higher eukaryote counterparts20,21,26. The unique feature of gw3’-RACE is its ability to monitor both tailed (short and long) and non-tailed reads in an RNA sample14.
We utilised gw3’-RACE to study uridylation in fission yeast. We observed that in wild-type cells, the main targets of uridylation were mRNAs, ncRNAs, and mitochondrially encoded RNAs (Fig. 1d). We identified two types of uridylation: uridylation of poly(A) tails and oligouridylation, which refers to uridylation of non-tailed RNAs. Uridylation of poly(A) tails was a widespread modification, found in ~ 20% of all detected tails, with uridine residues preferentially added to short poly(A) tails. On the other hand, mRNA oligouridylation is a rare modification, detected in wild-type cells in only about 0.1% of all tailed reads. However, oligouridylation was a dominant modification of mitochondrial RNA (mtRNA), where oligo(U) tails accounted for 20% of all reads mapping to mtRNA (Fig. 1d). This is the first report of uridylation of mitochondrial RNAs in fission yeast, although its functional significance remains a mystery. Two terminal uridyltransferases – Cid1 and Cid16 – were responsible for the two types of uridylation: Cid1 catalysed most of the poly(A) tail uridylation, while Cid16 added oligo(U) tails to non-tailed RNAs and was probably responsible for some uridylation of very short poly(A) tails (Fig. 3a, b and Supplementary Fig. 6d, e). Intriguingly, our preliminary data indicate that two types of uridylation also occur in human cells, with mRNAs being oligouridylated after deadenylation (Supplementary Fig. 9).
We found that the mean transcript tail length was the primary determinant of mRNA uridylation frequency, with shorter tails being uridylated more often (Fig. 2e), however, additional factors also appeared to be involved in establishing the level of uridylation. Interestingly, the group of most frequently uridylated mRNAs exhibited different gene category enrichments compared to the group of mRNAs with the shortest tails (Fig. 2f). Genes involved in general sugar and amino acid metabolism were among the most uridylated in wild-type fission yeast cells.
Our results suggest that the uridylation of poly(A) tails in fission yeast has a dual function. Firstly, it facilitated the binding of the Lsm1–7 complex, thereby promoting decapping and 5’ to 3’ mRNA decay27. Deletion of the lsm1 gene led to the global accumulation of mRNAs due to decapping inhibition and results in build-up of short poly(A) uridylated tails (8–14 nucleotides total length), which we considered substrates for the Lsm1–7 complex (Fig. 4e). Secondly, uridylation prevents excessive deadenylation which in turn protects mRNA from 3’ to 5’ exonucleolitic decay. This hypothesis is supported by several observations. Increased deadenylation was observed in the single cid1 deletion strain (Fig. 3e, f) and is enhanced by impairing of 3’ to 5’ decay pathways (Fig. 5d). Peak of short poly(A) uridylated tails observed in Δlsm1 strain is lost in Δcid1Δlsm1 strain, which coincides with extensive deadenylation (Fig. 4e) and accumulation of deadenylated mRNAs (Fig. 4g). The stabilising effect of uridylation in lsm1 deletion background is also supported by a positive correlation between uridylation frequency and accumulation of transcripts and proteins in the Δlsm1 strain (Fig. 4i, l). Moreover, we observed faster decay rates in the Δcid1Δlsm1 strain compared to the lsm1 deletion strain (Supplementary Fig. 6b, c).
Interestingly, the tail profile of some of the highly uridylated genes resembles that observed in strains with inhibited 5’ to 3’ decay, such as Δlsm1 strain (Fig. 2g). This suggests that for some genes, 5’ to 3’ decay rate is a turnover bottleneck and, for this group, uridylation of partially deadenylated transcripts may be especially important.
Inhibition of 3’ to 5’ decay pathways in strains with RNA 3’-ends left unprotected due to the deletion of cid1 and/or lsm128 led to increased oligouridylation (Fig. 5a, d). This suggests a build-up of non-adenylated mRNA substrates. We speculate that oligouridylation of de-adenylated mRNAs supports their decay in the 3’ to 5’ direction, however, from our data we cannot distinguish which of the 3’ to 5’ decay pathway depends on oligouridylation. Nonetheless, Dis3l2 has been shown to have a preference for uridylated RNAs in vitro and in vivo, while exosome-mediated 3’ to 5’ decay supported by the SKI complex is conserved in budding yeast, where uridylation is absent, and where it was shown to target non-tailed mRNAs29. Therefore, we suggest that in fission yeast, the Dis3l2 exonuclease targets oligo(U)-tailed mRNAs, while the SKI complex assists the exosome in removing non-tailed and short-tailed mRNAs. In such a scenario, an increase in oligouridylated mRNAs in the Δski2 deletion backgrounds could be a result of the accumulation of non-degraded exosome targets that are available for oligouridylation.
Based on our data, we propose a model of bulk mRNA decay that takes into account the dual role of uridylation in the process (Fig. 6). Under standard conditions, uridylation of short mRNA poly(A) tails by Cid1 helps to direct mRNA toward the 5’ to 3’ decay pathway by enhancing Lsm1–7 complex binding and protecting the 3’-end from extensive deadenylation. Transcripts that escape this pathway become extensively or completely deadenylated and are either removed by the exosome complex or further oligouridylated by Cid16 and degraded by the U-dependent Dis3l2 exonuclease. We hypothesise that uridylation protects mRNA from further shortening until a certain tail length, thereby favouring the shift of decay balance towards the 5’ to 3’ degradation pathway. However, below the critical threshold, it triggers 3’ to 5’ decay.
We propose a model of bulk mRNA decay that takes into account the dual role of uridylation in the process. Under standard conditions, uridylation of mRNA poly(A) tails by Cid1 helps to direct mRNA toward the 5’ to 3’ decay pathway by enhancing Lsm1–7 complex binding and protecting the 3’-end from extensive deadenylation. By providing 3’-end protection, uridylation represses the 3’ to 5’ decay pathways that target predominantly excessively deadenylated transcripts. Deadenylated mRNAs are removed by the exosome complex or can be oligouridylated by Cid16 TUTase which targets their degradation by Dis3l2 U-dependent exonuclease.
The main function of uridylation of poly(A) tails in fission yeast is, therefore, to support 5’ to 3’ decay and (concomitantly) to protect from 3’ to 5’ decay. Interestingly, the deletion of the ski2 gene in the Δcid1 background results in strong growth inhibition. The SKI complex is an important contributor to co-translational surveillance-decay pathways30. It is tempting to speculate that the higher contribution of 3’ to 5’ mRNA decay puts additional pressure on co-translational surveillance due to interference between translation and decay. It was previously suggested that the dominance of 5’ to 3’ mRNA decay in eukaryotic cytoplasm helps to avoid clashes between translation and decay since both processes act in the same direction along the mRNA31,32.
In conclusion, we provided evidence supporting the dual role of uridylation in triggering decay and protecting against excessive deadenylation, which helps route mRNA to the 5’ to 3’ decay pathway. Further research will be necessary to assess the exact contributions of differently directed decay in bulk degradation and to determine the extent to which uridylation controls this balance.
Methods
Yeast strains and media
Wild-type S. pombe 972 strains of two mating types: h- or h +s, were used as a control and to create all deletion mutants. Yeasts were grown in standard conditions and media (32 °C in yeast extract with supplements (YES), or in Edinburgh minimal medium (EMM).
Strains construction
For the preparation of single deletion strains standard homologues recombination based protocol was used. The pFA6A backbone plasmids were used to produce recombination cassettes bearing different antibiotic resistance33. For double and triple mutant construction, appropriate strains were crossed and after spore dissection, strains with the right allele combinations were selected based on antibiotic resistance. A list of yeast strains created in this study is available in the Supplementary File. The identity of strains was controlled by colony PCR. Supplementary Fig. 10 confirms depletion of genes of interest in 15 mRNA degradation mutants. Selected gw3’-RACE data visualisations for 15 mRNA degradation mutants are provided in Supplementary Figs. 7 and 8.
Growth assays
To compare the growth of different strains in liquid media changes in optical density (OD 600) were recorded using micro-bioreactor Bioscreen C. Cells from overnight pre-cultures were set to OD 600 of 0.1 and grown in 100-well plates in 100 μL volume. Optical density was measured every 15 min. Growth curves were extracted from the data using the Pyphe growth curves Python module4 and visualised using custom R scripts. Growth statistics were calculated using Growthcurver package34.
To compare yeast growth in solid media yeast cultures were grown in YES in standard conditions to OD 0.5, next cells were washed with PBS, and five-fold dilution series was prepared in a sterile 96-well plate. Dilutions were next spotted onto agar plates using a 96 pinner.
gw3’-RACE library preparation
Yeast RNA was extracted from exponentially grown cultures with hot phenol protocol followed by clean-up using Qiagen RNeasy Kit column with DNase I treatment. The rRNA depletion was achieved using a riboPOOL kit (siTOOLs). After depletion, RNA was cleaned-up using MiniElute Cleanup columns (Qiagen) and adenylated 3’-adaptor was ligated using T4 RNA ligase 2 (NEB). The reaction product was purified using MiniElute Cleanup (Qiagen) and reverse transcription was performed using Superscript III (Thermo). The product was purified using AMPure beads and second strand synthesis was performed like in ref. 19 using E.coli DNA polymerase, E.coli DNA ligase I and NEBNext Second Strand Synthesis Buffer. The reaction product was cleaned up using AMPure beads. Double-stranded DNA was fragmented using tagmentase (Tn5) loaded with i5 compatible adaptor. Tn5 enzyme was obtained as described in ref. 35. Products were cleaned up using AMPure beads. The Detailed protocol was included in the Supplementary Information file. The final library was PCR amplified using NEXTERA index primers. Detailed protocol, adaptors and primer sequences can be found in Supplementary Information. Libraries were sequenced using NOVA-seq equipment paired-end sequencing with 100nt read length. Raw sequencing data were deposited in the Sequence Read Archive SRA NCBI) under accession number PRJNA945074.
gw3’-RACE data processing
Sequencing data were filtered for read pairs where R2 contained a 3’-adaptor sequence ligated at the first steps of the library preparation (using text search and fastq_pair). Short reads and low-quality read pairs were filtered out using fastp36. Reads R1 and R2 were aligned separately to the genome using STAR aligner37, in the case of R2, settings allowed soft-clipping and keeping non-aligned reads in the final output (.sam). Next, using samtools (version 1.10)38 and bedtools (version 2.27.1)39 utilities, uniquely aligned reads were extracted from the R1 alignment file and matched to genomic features (genes) forming a.bed type file. Bed files were transformed into bigwig files for visualisation using the bedGraphToBigWig utility (version 4). In the next step, R1 reads names from created .bed file was matched with R2 read mates from the.sam file, custom table was created containing in each row R2 CIGAR string, read name and sequence as well as R1 mate matched feature and R1 and R2 alignment coordinates. These raw data tables were a base for tail analysis information about the tail existence, its length and type were then extracted from the tables using a custom Python (v3.9) script (https://github.com/igib-rna-tails/gw3-RACE_analysis). The script extracted tail sequence from R2 read based on CIGAR string and categorised tails as poly(A), poly(A)U, oligo(U), or other using text search (grep regular expressions). Noteworthy, sequencing longer homopolymers resulted in a drop of per base sequence quality for nucleotides behind poly(A) stretch (Supplementary Fig. 1d), which in turn prevented alignment of R2 reads with long untemplated tails (Supplementary Fig. 1e). If R2 was not aligned and therefore CIGAR was not available R2 3’-end sequence was scanned using text search to categorise the tail into one of the aforementioned four categories. Tails were strictly defined with no mismatches allowed. A and U tails of minimum 1 nucleotide and AU tails from 3 nt were searched. Uridine residues were only permitted at the 3’ termini in the case of poly(A) uridylated tails. The tails that did not meet the criteria were categorised into the ‘other’ tails subcategory. The script produced as output a.csv type table with the information necessary for downstream data analysis and visualisation and with sequence of extracted tail and UMI (tables are available here https://osf.io/wtb49/). Data was further de-duplicated using information of R1 mapping and UMI sequence. The structure of data pre-and post de-duplication is represented in Supplementary Fig. 11. Scripts sufficient to perform the analysis are included in the Supplementary Information file and pipeline schematics are provided in Supplementary Fig. 2. Some mapping and filtering statistics for gw3’-RACE samples used in this study are provided in Supplementary Fig. 12.
RNA-seq and differential expression analysis
RNA was extracted, and rRNA was depleted like in the gw3’-RACE protocol. Stranded RNA-seq libraries were next prepared using NEXTFLEX Rapid Directional RNA-Seq Kit (PerkinElmer). Libraries were sequenced using NOVA-seq equipment paired-end sequencing with 100nt read length. Sequencing data were filtered using fastp36, and aligned to the genome using STAR aligner version 2.7.3a37. Reads aligning to individual genes were counted using the htseq-count module40 and differential expression analysis was performed using the DESeq2 R package41 using the adaptive t prior shrinkage estimator from the apeglm package42. DESeq2 results tables are included in Supplementary Datasets 5–7 files.
Data analysis and visualisation
Final analysis and visualisation of the data were performed in R (version 4.0.4). Input for analysis consisted of tables deposited in OSF, and custom functions for data filtering and calculating tail-specific values per gene were prepared. A list of genes that were excluded from the analysis due to documented overlap with non-coding features like sn or snoRNAs (16 genes) and 10 coding genes that were excluded because of peculiar coverage patterns suggesting presence of non-coding RNAs or technical issues is provided in Supplementary Dataset S3.
Proteomics
Proteins were extracted from exponentially growing cells and subjected to reduction, alkylation, and tryptic digestions, as described previously43. Proteomic data was acquired using microflow chromatography and SWATH-MS on a 7600 instrument (Sciex)44. Proteoms were measured for 15 different fission yeast strains with mutated RNA decay machinery (see strain list in Supplementary Fig. 10), and at least 5 repeats were measured for each mutant. Raw data was processed using DIA-NN45. Differentially expressed proteins in Δlsm1 compared to wt strain were identified using the DEP R package46, analysis output can be found in Supplementary Dataset S8.
mRNA half-life measurements
Total RNA was extracted with the hot phenol extraction method. For northern blot analysis, total RNA (10 μg) was resolved in 1.1% agarose/6% formaldehyde gel and transferred by capillary elution to a nylon membrane (Amersham). Random primed probes were amplified using a DNA template with appropriate primers (Supplementary Dataset S2) and radioactively labelled with DecaLabel DNA Labelling Kit (Thermo Scientific) and [α−32P]ATP (Hartmann Analytics). The oligoribonucleotide probe (for 25 S) was radioactively labelled using PNK (Thermo Scientific) and [γ-32P]ATP (Hartmann Analytics). Membranes were hybridised overnight with radioactive probes in PerfectHyb buffer (Sigma-Aldrich), washed, analysed with PhosphorImager Typhoon FLA 9000 (GE Healthcare) and quantitated with ImageJ software. mRNA half-life was calculated by regression of the semi-logarithmic mRNA levels against time. 25 S rRNA was used for the normalisation of expression values. Experiments were performed in three independent biological replicates. Raw Northern blot pictures and calculated half-lifes are available in the OSF repository [https://osf.io/wtb49/].
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. Raw sequencing data were deposited in Sequence Read Archive (SRA, NCBI) under accession number PRJNA945074. Raw proteomics data are available via ProteomeXchange with the identifier PXD051392. Protein intensities calculated for individual samples are available in the OSF repository [https://osf.io/wbxe7/]. Tables with gw3’-RACE non-deduplicated data and extracted tail sequences, as well as genomic annotation files and other additional files, are available in the OSF repository [https://osf.io/wtb49/]. Raw Northern blot pictures and calculated half-lifes are available in the OSF repository [https://osf.io/wtb49/]. Source data are provided in this paper.
Code availability
Code for gw3’-RACE analysis of raw sequencing data is available in GitHub: https://github.com/igib-rna-tails/gw3-RACE_analysis alternativelly under https://doi.org/10.5281/zenodo.12207185. Code for post-processing is available in Supplementary methods and GitHub: https://github.com/igib-rna-tails/gw3-RACE_vizualization.
References
Pérez-Ortín, J. E., Tordera, V. & Chávez, S. Homeostasis in the Central Dogma of molecular biology: the importance of mRNA instability. RNA Biol. 16, 1659–1666 (2019).
Parker, R. RNA Degradation in saccharomyces cerevisae. Genetics 191, 671 (2012).
Passmore, L. A. & Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 23, 93–106 (2022).
Halbach, F., Reichelt, P., Rode, M. & Conti, E. The yeast ski complex: Crystal structure and rna channeling to the exosome complex. Cell 154, 814–826 (2013).
Anderson, J. S. J. & Parker, R. The 3’ to 5’ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3’ to 5’ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 (1998).
Dave, P. & Chao, J. A. Insights into mRNA degradation from single-molecule imaging in living cells. Curr. Opin. Struct. Biol. 65, 89–95 (2020).
Lim, J. et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365–1376 (2014).
Rissland, O. S. & Norbury, C. J. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623 (2009).
De Almeida C., Scheer H., Zuber H., Gagliardi D. RNA uridylation: a key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdiscip. Rev. 9, e1440 (2018).
Zigáčková D., Vaňáčová Š. The role of 3′ end uridylation in RNA metabolism and cellular physiology. Philos. Trans. R. Soc. B Biol. Sci. 373, https://doi.org/10.1098/rstb.2018.0171 (2018).
Scheer, H. et al. The TUTase URT1 connects decapping activators and prevents the accumulation of excessively deadenylated mRNAs to avoid siRNA biogenesis. Nat. Commun. 12, 1–17 (2021).
Zuber, H. et al. Uridylation and PABP cooperate to repair mRNA deadenylated ends in arabidopsis. Cell Rep. 14, 2707–2717 (2016).
Malecki, M. et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 32, 1842–1854 (2013).
Brouze, A., Krawczyk, P. S., Dziembowski, A. & Mroczek, S. Measuring the tail: Methods for poly(A) tail profiling. Wiley Interdiscip. Rev. 14, e1737 (2023).
Rodriguez-Lopez, M. et al. Functional profiling of long intergenic non-coding RNAs in fission yeast. Elife 11, e76000 (2022).
Preston, M. A. et al. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445 (2019).
Lipińska-Zubrycka, L., Grochowski, M., Bähler, J. & Małecki, M. Pervasive mRNA uridylation in fission yeast is catalysed by both Cid1 and Cid16 terminal uridyltransferases. PLoS ONE 18, e0285576 (2023).
Pisacane, P. & Halic, M. Tailing and degradation of Argonaute-bound small RNAs protect the genome from uncontrolled RNAi. Nat. Commun. 8, 15332 (2017).
Alpern, D. et al. BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 20, 71 (2019).
Wiener, D., Antebi, Y. & Schwartz, S. Decoupling of degradation from deadenylation reshapes poly(A) tail length in yeast meiosis. Nat. Struct. Mol. Biol. 28, 1038–1049 (2021).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: Genome-wide determination of poly(A) tail length and 3’ end modifications. Mol. Cell 53, 1044–1052 (2014).
Yi, H. et al. PABP Cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell 70, 1081–1088.e5 (2018).
Marguerat, S. et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 (2012).
Lima, S. A. et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 24, 1057–1063 (2017).
Tudek, A. et al. Global view on the metabolism of RNA poly(A) tails in yeast Saccharomyces cerevisiae. Nat. Commun. 12, 4951 (2021).
Song, M. G. & Kiledjian, M. 3’ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13, 2356–2365 (2007).
He, W. & Parker, R. The yeast cytoplasmic lsm1/pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158, 1445–1455 (2001).
Benard, L., Carroll, K., Valle, R. C. P. & Wickner, R. B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell Biol. 18, 2688–2696 (1998).
Bae, H. & Coller, J. Codon optimality-mediated mRNA degradation: Linking translational elongation to mRNA stability. Mol. Cell 82, 1467–1476 (2022).
Huch, S. & Nissan, T. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. 5, 747 (2014).
Heck, A. M. & Wilusz, J. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032839 (2018).
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Sprouffske, K. & Wagner, A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinforma. 17, 172 (2016).
Hennig, B. P. et al. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 Genes Genomes Genet. 8, 79–89 (2018).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Messner, C. B. et al. The proteomic landscape of genome-wide genetic perturbations. Cell 186, 2018–2034.e21 (2023).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Wang, Z. et al. High-throughput proteomics of nanogram-scale samples with Zeno SWATH MS. Elife 11, e83947 (2022).
Zhang, X. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018).
Eser, P. et al. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol. Syst. Biol. 12, 857 (2016).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Acknowledgements
We would like to thank David Elis for the critical reading and editing of the final version of the manuscript; Roman Szczęsny for the critical reading, fruitful discussions, and providing HeLa cells RNA material, Joanna Kufel for support and critical reading of the manuscript, Paweł Golik for support and providing budding yeast RNA material, Andrzej Dziembowski for advice, Jürg Bähler for advice and critical reading of the manuscript. Michael Mülleder and Anja Freiwald for help with proteomics data deposition. This research was funded by the Foundation for Polish Science FIRST TEAM grant (POIR.04.04.00-00-4316/17) awarded to Michał Małecki. NGS sequencing was performed at the University of Warsaw Centre of New Technologies Genomics Facility. Proteomics was performed in HT-MS | High Throughput Mass Spectrometry Core Facility at Charité Universitätsmedizin, Berlin, Germany.
Author information
Authors and Affiliations
Contributions
M.G. and L.L.-Z. performed majority of the laboratory work, initial data processing and analysis and participated in downstream analysis and manuscript preparation; StJ.T. and M.R. coordinated and performed mass spectrometry and proteomics data processing; A.G.-M. and M.Z.-P. performed Northern blot analysis and half-life calculations, B.J. prepared some of gw3’-RACE libraries; G.B. and S.Ś. purified tagmentase; S.Ś. and M.R. helped to edited final manuscript; M.M. conceptualised the project, organised funding, managed work, wrote manuscript, prepared figures and performed majority of downstream data analysis. A.G.-M. and M.Z.-P. contributed equally.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Eugene Valkov and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Grochowski, M., Lipińska-Zubrycka, L., Townsend, S. et al. Uridylation regulates mRNA decay directionality in fission yeast. Nat Commun 15, 8359 (2024). https://doi.org/10.1038/s41467-024-50824-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50824-w