Abstract
The nematode Caenorhabditis elegans is widely employed as a model organism to study basic biological mechanisms. However, transgenic C. elegans are generated by manual injection, which remains low-throughput and labor-intensive, limiting the scope of approaches benefitting from large-scale transgenesis. Here, we report a robotic microinjection system, integrating a microfluidic device capable of reliable worm immobilization, transfer, and rotation, for high-speed injection of C. elegans. The robotic system provides an injection speed 2-3 times faster than that of experts with 7–22 years of experience while maintaining comparable injection quality and only limited trials needed by users to become proficient. We further employ our system in a large-scale reverse genetic screen using multiplexed alternative splicing reporters, and find that the TDP-1 RNA-binding protein regulates alternative splicing of zoo-1 mRNA, which encodes variants of the zonula occludens tight junction proteins. With its high speed, high accuracy, and high efficiency in worm injection, this robotic system shows great potential for high-throughput transgenic studies of C. elegans.
Similar content being viewed by others
Introduction
With the recent advances of gene editing technology, it has become feasible to identify disease-relevant human gene variants through genotyping hundreds of thousands of patients1,2. To fully take advantage of this technology and rapidly probe gene functions, large-scale genetic modifications are required. To overcome the ethical, economic, and experimental limitations associated with the use of mammalian models for early-stage gene editing studies, the nematode worm Caenorhabditis elegans, a small model organism, is advantageous for studying gene functions relevant to human diseases due to its small size, transparent body, ease of culture, short life cycle, many homologs of human genes, and large mutant libraries3,4,5. In addition, C. elegans is also widely used as a model for discovering basic biological mechanisms. For instance, optogenetics6,7, multimodal sensory integration8,9, the role of small RNAs10, and many other biological research topics11,12,13 all have greatly benefited from studies on this small organism.
To generate mutant worm strains, different genetic vectors need to be delivered into the distal gonads of C. elegans. To date, the most efficient method to create these transgenic mutants is through manual injection by a human operator under a microscope14,15. This conventional manual approach has remained unchanged for decades. For manual injection, a human operator usually cannot sustain continuous operation for more than several hours due to the fatigue caused by delicate manipulation under the microscope16. A highly experienced operator may perform the manual injection at a speed of 2 ~ 3 min per worm, while a common operator will need much more time for injection of one worm17. Due to its inherent drawbacks such as labor-intensiveness, operation inconsistency, and low throughput, the manual injection has become a major technical bottleneck for large-scale genetic studies of C. elegans.
Robotic systems hold great potential to automate the microinjection process of C. elegans16; however, the high-precision injection of plasmid into a target organ inside the worm body still remains problematic due to the small worm body size, various structures with irregular shapes inside the worm body, and the random body orientation after immobilization (usually on an agar pad). In this regard, several worm manipulation techniques (e.g., immobilization and rotation) have been realized on microfluidic devices and high manipulation accuracy and consistency have been achieved thanks to the merits of microfluidics such as channel-sample size matching, precise and parallel fluid manipulation, and on-chip integration of various manipulation strategies18,19,20. Furthermore, the synergy of robotics and microfluidics has opened a route to the accurate, high-efficiency, and intelligent microinjection of C. elegans17,21. Two microfluidic devices have been developed and integrated with robotic systems for sequentially loading and immobilizing C. elegans for microinjection17,21. However, in both systems17,21, the body orientation of a C. elegans is random upon microfluidic immobilization, therefore limiting the precise microinjection of plasmid into the target body structure.
To achieve precise, high-speed microinjection of C. elegans, we report an automated robotic system with a computer-controlled microfluidic device, which can stably rotate and immobilize a worm body and precisely inject solutions into a specific organ (e.g., the distal gonad) of the worm body. The microfluidic device, consisting of a row of open-ended channels, leverages precise pressure control (via a pressure regulator) for sequential rotation and immobilization of single worms, and the robotic system takes full advantage of image processing and automation techniques to precisely control the motions of an injection pipette and the microfluidic device for automated worm injection. To automate pipette penetration during the injection process, a closed-loop control system based on visual feedback is developed. Through integration of the developed microfluidic device and robotic system, the precise and high-speed injection of a worm is consistently achieved.
To demonstrate the capabilities of this robotic system, we successfully inject different worm strains with plasmid DNA solutions and ribonucleoprotein (RNA-protein or RNP) complexes to create stably inheriting extrachromosomal arrays and to achieve heritable genome editing by clustered, regularly interspaced, short, palindromic repeat (CRISPR)/CRISPR-associated endonuclease 9 (Cas9). This robotic system can perform single gonad arm injection at an average speed of 44.5 s/worm, which includes the entire injection process of worm transfer, immobilization, rotation, injection, and release, and is ~2–3 times faster than the injection speed (~2–2.7 min/worm) of experts who have ~7–22 years’ experience. For the experiments with plasmid DNA injection, the average number of transgenic progeny among the first filial (F1) generation produced by robotic single gonad arm injection and double gonad arms injection are comparable to those from the manual injection by the experts. We also find that the robotic system enables precise microinjection of worm mutants with body sizes different from the wild-type (N2) strain and anesthetized worms with curved bodies. In addition, there is no obvious damage caused to the worm by the robotic injection protocol. Operators without expertise in robotics can quickly learn the usage of our system after a few trials and perform the injection with high success rate and excellent injection quality. We demonstrate the high speed, accuracy, and efficiency of our robotic system by performing transgenesis experiments and further demonstrate the utility of the system in a large-scale reverse genetic screen. These results prove that our system can facilitate high-throughput transgenic studies in C. elegans.
Results
Design of the robotic system for C. elegans microinjection
To establish a robotic system for precise and high-speed microinjection of C. elegans, two motorized micromanipulators were integrated into a regular inverted microscope [Fig. 1a]. The left micromanipulator (MP-285, Sutter Instrument) has three orthogonal motion axes with a positioning resolution of 40 nm and a motion range of 25 mm along each axis [calibration data shown in Supplementary Fig. 1]. The right micromanipulator (MX7600, Siskiyou) has three orthogonal and one diagonal motion axes with a step resolution of 40 nm and a motion range of 20 mm along each axis. An injection holder (HI-7, Narishige) with a capillary glass pipette (for C. elegans injection) was mounted on the left micromanipulator with a tilting angle of 15° [Fig. 1b] for the penetration of C. elegans and the delivery of solution into the distal gonad using a pneumatic microinjector (IM-300, Narishige). A microfluidic device [Fig. 1c and d] for C. elegans manipulation was mounted on the right micromanipulator with a tilting angle of 30° [Fig. 1b, Supplementary Fig. 2]. To facilitate microinjection of C. elegans, there exists an angle of ~65° [indicated by the α in Supplementary Fig. 2d] between the x-y coordinates of the left micromanipulator frame and the camera frame as this angle allows the injection pipette to travel with a long distance inside the distal gonad. A custom-made reservoir with three adjacent areas, for worm loading, rotation/injection, and recovery, was prepared by bonding a patterned polydimethylsiloxane (PDMS) layer to a 3” × 2” glass slide with a thin PDMS layer coated on its top side [Fig. 1b, Supplementary Fig. 2k]. A miniaturized pressure regulator consisting of two miniature solenoid valves (10–32 UNF Female, 12 V DC, McMaster-Carr) and two vacuum pumps (SFE 12 V Air Pump, Robotshop) was used to regulate the negative pressure in a precise and quick fashion inside the microchannels of the microfluidic device [Supplementary Fig. 2i and j]. The detailed procedures for the system construction can be found in Supplementary Fig. 3 [Supplementary Table 2].
a Three-dimensional model of the robotic system for microinjection of C. elegans. b An enlarged view showing the detailed components of the robotic system. c Schematic of the microfluidic device. d Microscopic image of leftmost portion of microfluidic device mounted on the right micromanipulator under a 4× objective. Image is representative of at least 10 microfluidic devices. e Schematics of the microinjection procedure: (i) pick-up, (ii) transfer, rotation, and immobilization, (iii) injection, and (iv) release of the target worm. f Microscopic images showing the main operation steps during microinjection, including: (i)-(ii) rotation, (iii) immobilization, (iv) objectives switching from 4× to 10×, (v)-(vi) worm body penetration, (vii) plasmid DNA solution deposition, and (viii) pipette withdrawal. Images are representative of at least 20 microinjections. g Schematics of worm model and transverse sections of anterior/posterior part of worm at different orientations showing the relative position between intestine (purple area), distal gonad (blue area with yellow circles), and proximal gonad (blue area without yellow circles): (i) transverse sections of anterior worm body at different orientations, (ii) transverse sections of posterior worm body at different orientations. It should be noted that distal/proximal gonads are transparent, and intestine is opaque. h Mechanical model of worm rotation under the action of hydrodynamic forces and frictional force. i Continuous worm rotation at different orientations. Images are representative of at least 20 worm rotations.
Microinjection procedure and vision-guided robotic control
To enable worm injection in a reliable and controllable fashion, worms were anesthetized by 0.6 mg mL−1 levamisole solution, and their bodies remained relatively straight during the entire worm injection process. Before each injection experiment, the custom-made reservoir was filled with M9 buffer and 5-6 worms were quickly transferred into each loading area of the custom-made reservoir by a glass Pasteur pipette.
In each microinjection experiment [Supplementary Fig. 4], a C. elegans was manipulated following an operation sequence of pick-up [Fig. 1e-(i)], transfer [Fig. 1e-(ii)], rotation, immobilization, injection [Fig. 1e-(iii)], and release [Fig. 1e-(iv)]. To begin, a single worm in the worm loading area was picked up by the microfluidic device (through applying a constant negative pressure to the device inlet) and quickly transferred to the injection area to avoid collision between the microfluidic device and the PDMS walls of the custom-made reservoir. The worm was then released and rotated to the desired orientation (see below for rotation strategy), as shown in Fig. 1f-(i) and 1f-(ii). Thereafter, the worm was immobilized again by the microfluidic device [Fig. 1f-(iii)], and the image position of the target gonad was selected via computer mouse clicking on the screen by an operator and fed back to a proportional-integral-derivative (PID) controller that automatically drives the right micromanipulator to bring the selected gonad area to the vertical center of the field of view (FOV) along its vertical axis [ym-axis labeled in Supplementary Fig. 2d and f] and autofocused under 10× objective through the variance computation in grey level among image pixels [Fig. 1f-(iv), Supplementary Fig. 5]. Then, the injection pipette was moved from its home position [Supplementary Fig. 6] to the FOV [Fig. 1f-(v)] and penetrated the worm body based on the user-identified gonad injection location obtained through computer mouse clicking and visual feedback [Fig. 1f-(vi), Supplementary Figs. 4 and 7]. Finally, the solution was injected into the gonad [Figs. 1f-(vi) and 1f-(vii), Supplementary Movie 1]. Once the injection was completed, the injection pipette was withdrawn from the worm body [Fig. 1f-(viii)] and returned to its home position, and the injected worm was transferred to and released in the recovery area [Fig. 1e-(iv), Supplementary Fig. 8]. The schematic workflow of the entire worm injection process is shown in Supplementary Fig. 4, and images showing the entire worm injection process can be seen in Supplementary Fig. 8 [Supplementary Movie 2]. There are two special cases of the worm injection in which some operation steps could be omitted. If the gonad of worm is clearly observed and exposed to the injection tip after transfer operation, the subsequent rotation and immobilization steps can be omitted, and the worm can be directly injected [Supplementary Movie 2]. In addition, if some of worms are loaded in the junction region of loading area and rotation/injection area of custom-made reservoir in which loaded worms are away from the PDMS walls of the custom-made reservoir, these worms can be directly rotated for the injection, and thus the process of pick up and transfer can be omitted [Supplementary Movie 3].
The set angle α [Supplementary Fig. 2d] between the injection needle and the immobilized worm body may vary slightly every time an injection pipette is mounted onto the left micromanipulator. Therefore, after an injection pipette is mounted, the coordinate transformation from the microscopic image frame to the left micromanipulator frame will be re-calibrated through affine transformation by the least square method. During the microinjection process, the user-identified injection position of the gonad coordinates of each worm in camera frame under 10× objective varies. To automate pipette penetration during the injection process, a closed-loop control algorithm based on visual feedback was developed to precisely control the movement of injection pipette [Supplementary Fig. 7]. The position of the injection pipette tip in the camera frame was extracted by a real-time image processing algorithm [Supplementary Fig. 7a] and then transformed to the coordinates of the micromanipulator to form the closed control loop [Supplementary Fig. 7b], forming a vision-guided, look-then-move system.
Microfluidic device design
The microfluidic device [Fig. 1c and d, Supplementary Fig. 9] includes a row of open-ended microfluidic channels, all connected to a computer-controlled pressure regulator, for robotic worm rotation and immobilization. The device was fabricated from PDMS through soft lithography22,23. Through microfluidic flow regulation and vision-guided robotic control, the worm rotation and immobilization were realized to facilitate the high-speed microinjection of C. elegans [Supplementary Movies 4–6]. The open-ended channels are all connected with a single device inlet in a bifurcation layout [Supplementary Fig. 9], which ensures equal fluidic resistances between their open ends and the device inlet and thus uniform sucking pressures at the open ends. To provide a sufficient negative pressure while preventing C. elegans from being sucked into the open-ended microchannels, these microchannels were designed to be 50 μm wide and 25 μm high. To seal these channels, a 14 μm-thick PDMS layer was bonded to the bottom side of the top PDMS layer [Fig. 1c, Supplementary Figs. 9 and 10]. Note that the microfluidic device was mounted on the right micromanipulator with a tilting angle of 30° [Fig. 1b]. Thus, a slope of 45° was cut at the side wall of the microfluidic device where the open ends of the microchannels are arranged [Fig. 1c, Supplementary Fig. 9]. This slope allows only the device edge with the open channels to contact the worm body during immobilization and also avoids any image shading of the worm body by the device edge in the entire process of worm manipulation. We further designed a custom-made pressure regulator [Supplementary Fig. 2i and j], which can rapidly provide a negative pressure to the microfluidic device inlet for worm immobilization and injection. The maximum negative pressure of the pressure regulator system is −12.87 PSI, which is sufficient for secure C. elegans immobilization during injection [Supplementary Fig. 2l]. The worm loading reservoir was prepared by bonding a patterned PDMS layer to a 3” × 2” glass slide with a thin PDMS layer coated on its top side [Fig. 1b, Supplementary Fig. 2k].
Robotic rotation, immobilization, and injection of C. elegans
To generate transgenic strains, solutions containing DNA, RNA, and/or protein must be delivered into the distal gonad, rather than the intestine, the body cavity, or the proximal gonad. In this case, either of the distal gonads at the anterior/posterior parts of the worm body should be precisely rotated to an orientation facing the injection needle, therefore allowing the needle tip to approach and penetrate the distal gonad for material delivery. The cross-section through the middle portion of the anterior/posterior parts of the worm body mainly consists of three structures: intestine, distal gonad, and proximal gonad, as schematically described in WormAtlas24 and Fig. 1g. In addition, the intestine occupies approximately half of the body cross-section area, and the distal gonad and the proximal gonad each take up one-quarter of the same cross-section, respectively, as shown in Fig. 1g-(i) and 1g-(ii). Figure 1g also schematically shows the relative position between the intestine (purple area), the distal gonad (blue area with yellow circles), and the proximal gonad (blue area without yellow circles) at different orientations if a worm is placed in the loading reservoir with its head facing outwards. For the transverse section of the anterior middle part [A-A in Fig. 1g], the distal gonad is located to the left side of proximal gonad when the anterior intestine is above the anterior proximal and distal gonad [Fig. 1g-(i): 0° orientation, Supplementary Fig. 11]. At this orientation, the posterior distal gonad (B-B in Fig. 1g) is located to the left side of proximal gonad and distributed above the posterior intestine [Fig. 1g-(ii): 0° orientation]. This is because the adult worm intestine shows a dextral handedness to its position along the length of the animal such that if anterior intestine is localized at the top side, the posterior intestine would be located at the bottom side25.
For injection at the anterior distal gonad, it would be better to rotate the worm to a body orientation between 270° and 360° [Fig. 1g-(i)], in which the injection pipette on the left-hand side can approach the anterior distal gonad [Fig. 1a]. Different from the anterior gonad injection, the worm should be rotated to a body orientation between 0° and 90° [Fig. 1g-(ii)] for injection at the posterior distal gonad. If a worm is placed with its tail facing outward, the cross-section is shown in Supplementary Fig. 12. Under brightfield microscopy, both the distal gonad and proximal gonad are transparent, but the intestine is relatively opaque. To monitor rotation angle of the worm body, we visually recognized the pattern of the transparent gonad under a brightfield microscope, as shown in Fig. 1f and i. Figure 1i-(ii) shows the worm after being rotated by 90°, indicated by the smallest size of the observed transparent area under the bright field mode (labeled by white arrow). After that, the worm was further rotated to 180° and 270°, where the size of bright area reached the maximum [Fig. 1i-(iii)] and minimum [Fig. 1i-(iv)], respectively.
As mentioned above, the worm rotation is required for successful injection. Here, we propose a simple but quite effective method for stable worm rotation. The microfluidic device was first connected with a plastic tube, and the plastic tube was then connected with the pressure regulator. The microfluidic device and the plastic tube from the inlet of microfluidic device to the part at highest point were filled with M9 medium by turning on the miniaturized pressure regulator. When there was no negative pressure applied to the microfluidic device, the M9 medium inside the microfluidic channels and the tube would slowly flow out of the microchannel open ends under the action of gravity. Once a worm was moved close to the open ends, it would be subjected to several forces [Fig. 1h], including the frictional force (Fpf) between the worm body and the substrate, and the drag force (Fd) and the lift force (Fl) caused by the laminar fluid flow out of the channel open ends [see Supplementary Methods and Supplementary Fig. 13]. The closer the worm body is to the channel open ends, the larger the drag force will be. It was found when the worm body was moved close to the open ends of the microfluidic device, the drag force (Fd) would finally counterbalance the frictional force (Fpf) to prevent the worm body from being in contact with the channel open ends [Fig. 1h]. As shown in Fig. 1i, during worm rotation there is always a gap between the worm body and the open-end edge of microfluidic device, and this gap would keep consistent during the rotation process as the worm body is finally rotated within the laminar region [see Supplementary Methods, Supplementary Fig. 13, and Fig. 1h].
In the vertical direction, the force of gravity on the worm is equal to combination of the lift force (Fl), the buoyance force (Fb), and the normal force (N) applied to the worm body by the petri dish. The height of microfluidic channel center at the open ends of microfluidic device was calculated to be 36.2 μm which is higher than that of worm body center (~25 μm). Due to the 45° slope and relatively large height of microfluidic channel at the open ends of microfluidic device, the hydrodynamic forces from fluid flowing out of the channel open ends would mainly act on the top surface of worm body [Supplementary Fig. 13b]. As the frictional force acts on the bottom side of worm body and hydrodynamic forces are mainly exerted on the top surface of worm body, a net rotational torque in counterclockwise direction will be thus exerted on the worm body [Fig. 1h]. Under the action of rotational torque generated from those forces, a worm can be reliably rotated [Fig. 1i, Supplementary Fig. 13, Supplementary Movies 4–6]. If the motorized stage was controlled to move forward in a relatively large speed without a stoppage, the worm body under the action of large lift force for a long duration would be lifted. A stoppage of the motorized stage for a certain duration after forward movement would enable the oriented worm body to rest on the substrate after rotation and always to be in focus. To obtain the consistent and accurate worm rotation [Fig. 1i], the motorized microscope stage holding the custom-made reservoir was controlled to move forward for a predetermined duration followed by stoppage for another duration.
Once a worm was rotated to the desired orientation, a negative pressure of −12.87 PSI was applied to immobilize the worm body. Supplementary Fig. 14a shows the sequential operations of worm including rotation, immobilization, and release. One can see that both anterior and posterior gonads of immobilized worm are exposed after rotation, shown in Supplementary Figs. 14a-(iii) and 14a-(vi), respectively, which enables both gonads to be injected via this reliable worm rotation method [Supplementary Fig. 14b]. It should be noted that the worm immobilization after rotation does not alter the worm body orientation (the transparent area, labeled by the white arrow in Supplementary Fig. 14a-(ii) and 14a-(iii), remains at the same orientation). In some cases, certain portion of the worm body was curved after anesthesia. Such a worm body can still be reliably rotated and immobilized for subsequent injection [Supplementary Fig. 15, Supplementary Movie 3], which demonstrates the robustness of the developed rotation method. In combination with the worm anesthetization and the contactless rotation method, mutant strain displaying roller phenotype and dumpy phenotype can be stably rotated to different orientations and securely immobilized thereafter, which further highlights the reliability of the worm rotation method, as shown in Supplementary Fig. 16. We also found that mutant animals with smaller body sizes than that of the standard N2 strain, even if not totally anesthetized, can still be effectively rotated for the injection thereafter [Supplementary Fig. 17 and Supplementary Movie 7].
As mentioned above, the tube from the inlet of the microfluidic device to the part at the highest point was filled with M9 medium at first. In addition to the movement of microscope stage, the maximum height of the tube [indicated by the h in Supplementary Fig. 2e] plays an important role for reliable worm rotation which relies on the hydrodynamic forces generated by fluid flowing out of the tube through the microchannel open ends by gravity. Thus, we conducted a systematic study on the effect of four operation parameters on the continuous worm rotation speed [results shown in Supplementary Fig. 18]: the forward movement speed, the forward movement duration, the rest duration of the microscope stage during worm rotation, and the maximum height of the tube connecting the microfluidic device to the pressure regulator above the substrate [Supplementary Fig. 18c–f]. Based on the experimental results [see Supplementary Methods and Supplementary Fig. 18], four operation parameters were finally optimized to be 250 μm/s (forward movement speed), 300 ms (the forward movement duration), 200 ms (the rest duration of the microscope stage during worm rotation), and 9 cm (the maximum height of tube above the substrate); thus, fast and reliable worm rotation was obtained with negligible drift along the body length, as shown in Fig. 1i.
Another important procedure of C. elegans injection is the worm body penetration. In the manual injection, after the pipette tip indents the worm body with a certain level of deformation, the operator usually taps the microscope stage gently to vibrate the pipette tip to facilitate worm body penetration. In robotic cell injection, a piezo-drill has been adopted to facilitate the cell membrane penetration26,27,28, which is based on a similar mechanism of vibrating the pipette tip. Here, we propose an operation protocol including three separate movement steps of the pipette tip for pipette tip positioning, worm indentation, and penetration, respectively, to realize easy worm body penetration [see Supplementary Methods and Supplementary Fig. 19]. The first movement step for pipette tip positioning relies on the developed closed-loop control system based on visual feedback, and the penetration of worm body counts on the inertial force from the step displacement of injection needle in the last movement step and worm indentation obtained from the second movement of pipette tip. With the optimization of three parameters including the pipette tip size, the movement duration of the pipette in the second movement step, and the step displacement of the pipette tip in the last movement step [see Supplementary Methods and Supplementary Fig. 19], a penetration rate of 100% was achieved. Note that most of the pipette tip size we used in our experiments except the injection of Cas9 ribonucleoprotein complex was in the range of 0.7–1 μm. As shown in Supplementary Fig. 20, there is no obvious damage caused to the worm by the developed worm injection protocol.
In summary, there are several advantages of the developed worm rotation method. First, the frictional force and hydrodynamic forces [Supplementary Fig. 13] together induce the rotational torque for contactless, precise, and stable rotation of a C. elegans. This rotation mechanism is independent of the worm body size and curvature, and the level of anesthesia. With its contactless nature and independence of body size/curvature, the developed worm rotation method allows the stable rotation of different mutant strains with varying body sizes or harboring resistance to the levamisole treatment. By adjusting the operation parameters of the motorized stage and the pressure regulator, we can rotate a worm in a continuous fashion. In addition, the hydrodynamic forces caused by the fluid flow out of the channel open ends keep a consistent gap between the worm body and the microfluidic device edge during rotation; thus, the transparent gonad inside the worm body can be clearly observed under the brightfield microscopy, which provides accurate visual feedback for controlling worm orientation. Moreover, the worm rotation method is simple and reliable and can be readily adopted for other applications such as high-resolution fluorescence imaging of C. elegans and other nematode species at different body orientations18.
Generation of transgenic strains by injecting plasmid DNA
To demonstrate the capability of the developed robotic system to generate transgenic C. elegans strains, we injected a plasmid DNA solution (2.5 ng/μL pCFJ90) that, when successfully injected into the gonad of an adult mother, can generate transgenic progeny expressing the fluorescent protein mCherry in the worm’s pharynx (a reporter gene driven by the myo-2 promoter), as shown in Fig. 2a. We first quantified the injection speed of our robotic system on animals injected in only one gonad arm, (single gonad arm injection). The robotic system can perform single gonad arm injection at an average speed of 44.5 s/worm (N = 30), which is ~2–3 times faster than those of manual single gonad arm injections by experts who have ~7–22 years of experience (~2–2.7 min/worm, N = 11–12) and ~2–4 times faster than the speed of proficient operators with 0.5–2 years of experience (~2–4 mins/worm, N = 12–14), with the latter group performing the majority of injections in a typical worm lab.
a Generation of transgenic worms by the robotic injection. Images are representative of at least 150 transgenic worms from microinjections. b Calibration curve of the average transgenic F1 progeny vs. volume of injected plasmid DNA solution (number of injected worms: N = 10 for the injected volume of 50 pL, 100 pL, 300 pL, 400 pL; N = 11 for the injected volume of 200 pL, 500 pL; mean ± s.d.). c Number of transgenic F1 progeny obtained from robotic injection with/without the rotation procedure (number of independent injections: N = 10, 12, 11, 12 for single gonad injections w/wo rotation and double gonad injections w/wo rotation, respectively; mean ± s.d.). Two-sided t-test: single gonad arm injection pw/wo rotation = 0.00189; double gonad arm injection pw/wo rotation = 0.00019. **p < 0.005. d Number of transgenic F1 progeny obtained from robotic injection and manual injections: (i) comparison between robotic injection and manual injections of three experts with ~7–22 years’ experience (experts A, B, and C) (number of independent injections: N = 10, 11, 11, 12, 11, 11, 12 for single gonad injection by robotic system, experts A, B, C, and double gonad injections by robotic system, experts A, C, respectively; mean ± s.d.), (ii) comparison between robotic injection and manual injections of six proficient operators (operators D, E, F, G, H, and I) (number of independent injections: N = 10, 12, 12, 12, 12, 12, 14, 11, 12, 12 12, 12, 14, 14 for single gonad injection and double gonad injections by robotic system and six proficient operators, respectively; mean ± s.d.). Two-sided t-test: *p < 0.05, **p < 0.005, and ns (not significant): p > 0.05. e Robotic single gonad injections by five operators (operators E, J, K, L, M): (i) success rate of robotic injections when operators performed robotic injections for four trials, (ii) comparison between the control group and robotic injection results obtained from the fourth trial of five operators (number of independent robotic injections: N = 12, 11, 10, 10, 10, 10 for five operators and control group, respectively; mean ± s.d.). Two-sided t-test: ns (not significant): p > 0.05. f Robotic injection result of mutant strain (VC4040) which has small body diameter: (i) images of young adult worm from mutant strain-VC4040 showing it has a small diameter (~38 μm). Images are representative of VC4040 from which 100% of animals express GFP signals in the pharynx, (ii) images of transgenic progeny obtained from the successful robotic injection of plasmid pCFJ90 into the distal gonad of young adult worms from mutant strain-VC4040. Images are representative of at least 20 transgenic progeny obtained from the successful robotic injection of plasmid pCFJ90 into VC4040. Source data are provided as a Source Data file.
Besides the single gonad arm injection, the efficiency of some applications such as CRISPR/Cas9-mediated genome editing is improved by double gonad arm injection, in which the genetic materials are injected into both gonad arms through two rounds of worm body penetration. Using the robotic microinjection system, both single gonad arm injection and double gonad arms injection were successfully achieved [Fig. 1f, Supplementary Fig. 13]. We assessed the performance of our robotic system on creating transgenic animals through single gonad arm injection (N = 119) and double gonad arm injection (N = 49). Two performance metrics were quantified, including the success rate of material delivery into the distal gonad and the mean number of transgenic progeny per injected adult. Importantly, transgenic F1 progeny (the first set of transgenic offspring from injected worms) were observed from 104/119 (87.39%) of the single gonad arm-injected parent worms and 47/49 (95.9%) of the double gonad arms-injected parent worms, indicating a high success rate of delivering DNA to the distal gonad which is comparable to the success rates of the expert A (single gonad arm injection: 100%, N = 11; double gonad arms injection: 100%, N = 11), the expert B (single gonad arm injection: 100%, N = 11), and the expert C (single gonad arm injection: 91.7%, N = 12; double gonad arms injection: 100%, N = 12).
Besides the success rate of material delivery, the average number of transgenic progeny from each injected parent also indicates the quality of robotic worm injection. In conventional manual injection, the injected volume of plasmid solution to generate genetic strains largely relies on the operator’s experience. With the developed robotic system, we accurately studied the effect of plasmid DNA volume on the transformation efficiency. Different volumes of pCFJ90 plasmid solution in the range of 50–500 pL were injected into the single gonads of young adult worms (N2), and the resultant average number of transgenic F1 progeny was recorded [Fig. 2b]. Single-factor ANOVA of the data shows significant difference (p = 0.0032) among the average number of transgenic F1 progeny with different injection volumes. One can see that the average number of transgenic F1 progeny increases with the injection volume and reaches its maximum (40.9 ± 26.5) at the injection volume of 300 pL. Thus, 300 pL was determined as the optimal injection volume for all the subsequent experiments.
With the optimal injection volume, the average number of transgenic F1 progeny produced through the robotic single gonad arm injection and the robotic double gonad arms injection were quantified to be 40.9 per injected worm and 84.5 per injected worm, respectively [Fig. 2c]. To demonstrate that the rotation is essential for highly efficient and successful worm injection, comparisons of the success rate and average number of transgenic progeny from each injected parent were made between the robotic injections with/without rotation procedure. As shown in Fig. 2c, the success rate from the robotic injections in which rotation was omitted (single gonad arm injection without rotation: 25%, N = 12; double gonad arms injection: 41.7%, N = 12) was much lower than those from the rotation-based worm injections. Also, compared to the rotation-based robotic injections, there existed a significant decrease in the average number of transgenic F1 progeny from each injected parent in which the rotation procedure was not involved (robotic injection without rotation: 4.5 transgenic F1 progeny/single gonad injected worm; 5.3 transgenic F1 progeny/double gonad injected worm; single gonad arm injection t-test: pw/wo rotation = 0.00189; double gonad arm injection t-test: pw/wo rotation = 0.00019). These results further indicate that the rotation is crucial for improving the success rate and quality of worm injection.
Also, as shown in Fig. 2d-(i), the average number of transgenic F1 progeny from the robotic single gonad arm injection shows no significant difference (t-test: pA, robot = 0.21; pB, robot = 0.40; pC, robot = 0.50) compared to the data produced by the experts with ~7–22 years of experience. There also existed no significant change between the average number of transgenic F1 progeny from the robotic double gonad arm injection and the data obtained from the expert A (t-test: pA, robot = 0.46; pC, robot = 0.15). These results indicate that the performance of robotic injection is comparable to that of the manual injections by the experts with ~7–22 years of experience. As shown in Fig. 2d-(ii), when compared to the manual injection from six proficient operators (operators D, E, F, G, H, and I) with ~0.5–2 years of experience, the robotic double gonad arms injection shows a significantly higher average number of transgenic F1 progeny than the results of manual double gonad arms injection from all operators (operator D: 8.5 transgenic F1 progeny/injected worm; operator E: 24.7 transgenic F1 progeny/injected worm; operator F: 32.9 transgenic F1 progeny/injected worm; operator G: 5 transgenic F1 progeny/injected worm; operator H: 26.4 transgenic F1 progeny/injected worm; operator I: 11.5 transgenic F1 progeny/injected worm; t-test: pD, robot = 0.00026; pE, robot = 0.0017; pF, robot = 0.0019; pG, robot = 0.00018; pH, robot = 0.0022; pI, robot = 0.00040). The average number of transgenic F1 progeny from manual single gonad arm injection by 5 proficient operators (D, F, G, H, and I) was significantly lower (operator D: 9.1 transgenic F1 progeny/injected worm; operator F: 18.17 transgenic F1 progeny/injected worm; operator G: 3 transgenic F1 progeny/injected worm; operator H: 8.6 transgenic F1 progeny/injected worm; operator I: 5.6 transgenic F1 progeny/injected worm; t-test: pD, robot = 0.0043; pF, robot = 0.029; pG, robot = 0.0013; pH, robot = 0.0037; pI, robot = 0.0040) than the robotic single gonad arm injection (40.9 transgenic F1 progeny/injected worm). These results further indicate that performance of robotic injection is much better than that of the most manual injections by proficient operators with ~0.5–2 years of experience, who carry out the majority of injections in a typical worm lab.
To demonstrate that the robotic system can be easily adopted by biological researchers (who have no expertise in robotics), we invited five graduate students, one skilled at the manual injection of C. elegans (operator E) and the other four with no prior experience in C. elegans injection (operators J, K, L, and M), to perform single gonad arm injection using the robotic system. The average number of transgenic F1 progeny from the robotic single gonad (40.9 transgenic F1 progeny/injected worm) was defined as control group for the following comparison. At first, all operators performed robotic injections over three trials and injected 10-12 worms per trial. As shown in Fig. 2e-(i), the success rate of robotic single gonad arm injection generally improved with each additional trial. Finally, the success rate of the fourth trail performed by each operator ranges from 80% to 90.9%. Also, the number of transgenic progeny for the fourth trial was recorded, as shown in Fig. 2e-(ii). By the fourth trial, the average number of transgenic progeny obtained from five operators (operators E, J, K, L, and M) show no significant difference from the control group (t-test: pE, control = 0.15; pJ, control = 0.12; pK, control = 0.073; pL, control = 0.35; pM, control = 0.12).
Taken together, these results indicate that our robotic system provides comparable success rate of material delivery into the distal gonad and the mean number of transgenic progeny per injected adult to manual injection of experts with ~7–22 years of experience, but significantly increases the speed of microinjection while minimizing effort, input, and fatigue by the user, which allows consistent and high-throughput injection. Different from the conventional manual injection method which requires trainees to spend several weeks to months to learn this technique and provides poor injection performance16, operators without expertise in robotics can quickly learn the usage of the developed robotic system after a few trials and perform the injection with high success rate and high injection quality.
In many applications used in our laboratories and others such as genetic interaction profiling29, alternative splicing reporter expression30, and genetic rescue31,32, plasmid DNA is required to be injected into mutant strains. The average diameter of a wild-type young adult worm is ~53.6 μm (N = 10). However, for some mutant strains, the diameter of a young adult worm is much smaller. To verify that the robotic system can be employed for the microinjection of mutant strains with smaller body sizes, young adult worms from the mutant strain-VC4040 (Genotype:F32B4.4(gk5114)[loxP + myo-2p::GFP::unc-54 3′UTR +rps-27p::neoR::unc-54 3′ UTR + loxP]))33, the average diameter of which is ~37.5 μm (N = 17), was selected for the robotic injection of the plasmid DNA pCFJ90 (2.5 ng/μL). Wild-type adult hermaphrodites typically produce ~300 progeny. However, the mutant strain (VC4040) is unhealthy as each adult worm produces a quite low number of progeny (on average 34.57 progeny per worm, N = 7). This mutant strain expresses green fluorescent protein (GFP) in the pharynx, as shown in Fig. 2f-(i). To securely immobilize the young adult worms from the mutant strain (VC4040) while avoiding these worms being sucked into microchannels, the negative pressure was reduced to −11.20 PSI for the successful injection. After successful robotic injection of plasmid solution into young adult worms, mCherry signal was observed in the pharynx of the transgenic progeny [Fig. 2f-(ii)]. Finally, transgenic F1 progeny were observed from 9/12 (75%) of the double gonad-injected parent worms from the VC4040, and the average number of transgenic F1 progeny per injected mutant worm was 2.9. The relatively low success rate and low average number of transgenic F1 progeny per injected mutant worm likely resulted from its unhealthy state and low brood size. All in all, these results indicate that our system can be easily accommodated to produce transgenic progeny from different mutant strains of varying body sizes.
When plasmid DNA solution is injected into the worm gonad, the DNA typically forms extrachromosomal arrays in the cells of progeny34. These extrachromosomal arrays are usually unstable during cell division but can become inheritable. Thus, most of the transgenic F1 progeny are transient and only a small fraction of transgenic F1 progeny will transmit stable transgenes arrays to the F2 generation34. To quantify the transgenesis efficiency using the developed robotic system, a mixed plasmid DNA solution (Pmyo-2::mCherry + Prgef-1::unc-16_minigene_splicing_reporter) was injected into the gonads of young adult worms (N2) in two independent trials (~15 worms injected for each trial). As the injected parent worms in each trial produced hundreds of transgenic F1 progeny, it is time-consuming to isolate and separately place each transgenic F1 progeny on a standard growth plate for following the inheritance pattern. In this case, we randomly selected 84 transgenic F1 progeny from each trial and placed each of them on a standard growth plate. An average percentage of 8.3% of transgenic F1 progeny transmitted the transgene to the second filial (F2), as shown in Table 1. This is a reasonable transmission rate from the extrachromosomal array bearing transgenic F1 progeny to the F2 generations as it is comparable to the reported rate of ~2–15% of transgenic F1 progeny transmitting extrachromosomal arrays to F2 generation35. These experiments further demonstrated the efficiencies of the developed robotic system in transgenic strain generation.
Generation of recombinant animals by CRISPR/Cas9 mediated genome editing
CRISPR RNAs and Cas9 protein have been widely used to create custom mutations and transgene insertions in C. elegans36,37. This revolutionary genome editing technology enables researchers to generate double-strand breaks at a target genomic locus by using Cas9 complexed with sgRNAs38. Upon cleavage by Cas9, the double-strand breaks at the target locus can undergo nonhomologous end joining (NHEJ) repair or homology-directed repair (HDR). Compared to the NHEJ, HDR allows for the precise editing of the genome by including a repair template, but at lower frequency, which is a good option for us to further test the performance of the robotic system.
To demonstrate that our robotic system can be employed to effectively generate recombinant worms using CRISPR/Cas9 genome editing techniques, we injected the CRISPR/Cas9 plasmid DNA solution (Pmyo-2::mCherry + Pmyo-3::mCherry + Peft-3::Cas9_SV40_NLS_tbb-2_UTR + pU6_klp-12_sgRNA + Pmyo-2::GFP_Prps-27::neoR_klp-12_disruption_repair_template)15 into both gonads of young adult worms. This CRISPR/Cas9-mediated genome editing is based on HDR wherein the repair template (Pmyo-2::GFP_Prps-27::neoR_klp-12_disruption_repair_template) disrupts the klp-12 gene and contains a dual-marker cassette, as shown in Fig. 3a, consisting of a GFP reporter expressed in the pharynx (Pmyo-2::GFP::unc-54 3′UTR) and a neomycin resistance (neoR) transgene (Prps-27::neoR::unc-54 3′UTR), which enables rapid identification of the rare recombinant worms after the microinjection. Detailed procedures for transgenic worm selection and screening can be found in Supplementary Fig. 21. To quantify the efficiency of plasmid-based CRISPR/Cas9 genome editing in C. elegans using the robotic system, we injected 30-48 worms each in three independent trials and successfully obtained the recombinant animals which survived G418 selection and possessed uniform GFP signal in the pharynx as well as absence of mCherry expression [Fig. 3b]. As seen in Table 2, from each of three independent injections (Exp. 1–3), we isolated two independent lines, for a total of six recombinant strains. As the plasmid-based CRISPR/Cas9 genome editing is a complex process which includes multiple steps such as transportation of plasmid DNA into the nucleus, transcription, and translation39, the editing is inefficient and the uncontrollable expression levels of the sgRNA and the Cas enzyme would lead to variable editing efficiencies across experiments. Operators D and F were invited to inject 30 worms with the same CRISPR/Cas9 plasmid DNA, and the insertion frequency was obtained to be 0% (0/30) and 3.3% (1/30), respectively. Thus, the obtained insertion frequency of 5.2 ± 1.4% is a reasonable efficiency for the plasmid-based CRISPR/Cas9 genome editing with a repair template when compared to the previously reported insertion frequency of 7.7–15% at the klp-12 locus from the manual injection performed by the expert A mentioned before14,15 and the insertion frequency of 0–3.3% obtained by the proficient operators. Although the GFP signal can be used as a marker to verify the insertion of dual-marker repair template, there exists the possibility of incomplete editing at the klp-12 locus of interest or off-target editing events for the plasmid-based CRISPR/Cas9 genome editing, the latter of which means that undesired insertion at other genomic loci may occur. To test for these possibilities in our recombinant worms, we genotyped each recombinant strain to confirm the correct insertion site. Our genotyping assays confirmed that 5/6 of the independent lines had the dual-marker cassette inserted in the correct location, giving amplicons that were specific to the upstream (us) and downstream (ds) insertion sites from recombinant animals [Fig. 3c, Table 2]. Only one line we recovered (Experiment 3, line 1) failed to insert correctly. However, this result is consistent with previous observations when performing manual injection by the expert A and is a reflection of the CRISPR editing and recombination process in the worm itself rather than a consequence of the robotic microinjection platform.
a Dual-marker repair template for CRISPR/Cas9 plasmid DNA-mediated genome editing. Purple boxes, green box, yellow box, and red triangles represent appropriate homology arms for HDR, a pharyngeal GFP marker transgene, a neoR transgene, and loxP sites, respectively. b Visual strategy for identifying the recombinants obtained from the injection of CRISPR/Cas9 plasmid DNA solution into wild-type worm: (i) captured image of the recombinant under the brightfield mode, (ii) uniform GFP expression in the pharynx of recombinants, (iii) loss of mCherry expression in the recombinant, (iv) only uniform GFP expression in the recombinant demonstrating integration of the repair template and loss of the extrachromosomal array. Images are representative of recombinant worm generated via CRISPR/Cas9 plasmid DNA-mediated genome editing from three independent batch of injections. c Genotyping results at the klp-12 gene locus after CRISPR/Cas9 genome editing experiments using robotic microinjection platform. Images are representative of genotyping results of 6 lines from three independent batches of injections. d Visual strategy for identifying the recombinants obtained from the RNP complex injection: (i) mutant strain-JAC644 with GFP signal expressed in pharynx was chosen for the RNP complex injection, Image is representative of JAC644 from which 100% of animals express GFP signals in the pharynx, which was selected for the injection. (ii) recombinant worm which survived the G418 selection and possessed a uniform mCherry signal in the pharynx was successfully obtained by injecting the RNP solution into mutant (JAC644). Image is representative of recombinant worm generated via RNP-mediated CRISPR genome editing from one batch of injections. Source data are provided as a Source Data file.
Another efficient method to realize the CRISPR/Cas9 mediated genome editing is to inject a pre-formed complex of recombinant Cas enzymes and chemically synthesized sgRNAs directly, which is also known as a Cas9/sgRNA RNP complex. Compared to the CRISPR/Cas9 plasmid-mediated genome editing, the introduced RNP complex can rapidly start CRISPR genome editing as it bypasses many steps including transportation of plasmid DNA into the nucleus, transcription, translation, all of which are required for the CRISPR/Cas9 plasmid-based genome editing40. However, the pre-formed RNP complex of sgRNAs and protein can be prone to aggregation, which makes the injection pipette more easily clogged when compared to the plasmid DNA. To avoid the problem of frequent clogging for the injection of the RNP complex, the pipette tip size should be sufficiently large. To test the capability of our robotic microinjection system using Cas9/sgRNA RNP complex-mediated genome editing, we chose the mutant strain JAC644 (Genotype: sup-46(csb40)[loxP + myo-2p::GFP::unc-54 3′ UTR + rps-27p::neoR::unc-54 3′ UTR + loxP]), in which GFP is expressed in the pharynx, as shown in Fig. 3d-(i). A solution containing RNP complex targeting sequences in the vicinity of the GFP open reading frame and linear double-strand DNA template encoding mCherry and a neomycin resistance (neoR) transgene plus 35 bases of flanking homology was injected into both gonads of JAC644. If successful, genome editing would replace GFP sequence with the mCherry open reading frame. Due to the easy aggregation of proteins and sgRNAs, pipettes with tip size larger than 1.5 μm were employed in the injection of RNP complex while the negative pressure of −12.87 PSI was still sufficient to securely immobilize C. elegans for the injection with large injection tip. Finally, we obtained the recombinant progeny which survived the G418 selection and possessed a uniform mCherry signal in the pharynx, as shown in Fig. 3d-(ii). The CRISPR/Cas9 mediated genome editing experiments demonstrate that our robotic system can accommodate a variety of injection solutions, including CRISPR/Cas9 plasmids and RNP complexes, for the worm injection.
Large-scale reverse genetic screen for regulators of alternative splicing
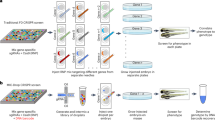
Alternative splicing is a crucial mechanism which significantly increases the complexity of gene expression41,42. However, regulation of alternative splicing is a complicated process, and it is still not completely understood42,43. Thus, to discover the mechanisms governing alternative splicing, large-scale reverse genetic screens employing transgenic mutant C. elegans can complement work in other model systems. As demonstrated above, our robotic system provides excellent injection performance in terms of injection speed and success rate, thus reducing the barrier to large-scale reverse genetic screens using available mutant strains generated by the C. elegans community. As such, we used our injection system to design a multiplexed reporter screen to identify candidate regulators of alternative splicing in neurons and intestinal cells [Fig. 4]. We previously developed a series of two-color fluorescent frameshift reporters that recapitulate tissue-specific alternative splicing patterns of endogenous alternative exons30. Normally, when these reporters are injected individually, the inclusion or skipping of an alternative exon can shift between two reading frames and translation of mCherry or GFP, respectively [Fig. 4a]. Thus, fluorescence can be visualized by microscopy as an indirect readout of alternative splicing, with GFP and mCherry signals providing relative measurements of exon skipping or exon inclusion, respectively30. We also use semi-quantitative RT-PCR as a readout to more directly assess reporter alternative splicing patterns30 [Fig. 4a]. We thus reasoned that we could inject a cocktail of multiple reporters simultaneously into wild type and mutant strains of interest, utilize reporter fluorescent signal to select for stable extrachromosomal array carrying animals, and then screen for effects on individual reporter splicing patterns by RT-PCR as a readout [Fig. 4b]. We prepared a cocktail of four splicing minigene reporters expressed specifically in neurons (Prgef-1::zoo-1 exon 9, Prgef-1::hlb-1 exon 12, Prgef-1::ampd-1 exon 15) or in the intestine (Pges-1::fhod-1 exon 9). These reporters were selected on the basis of displaying robust exon skipping in neurons but inclusion in other tissues (zoo-1, hlb-1, and ampd-1 reporters) or robust exon inclusion in the intestine compared to other tissues (fhod-1 reporter)30. This two-color fluorescent reporter cocktail was injected into wild-type animals and 48 mutant strains harboring deletion or loss of function alleles for different genes encoding known RNA binding proteins [Supplementary Data 1]. These mutants were selected because they represented all available alleles of RNA-binding protein genes available at the Caenorhabditis Genetics Stock Center (CGC) which could be maintained as homozygotes. After injection, we isolated and maintained stable extrachromosomal array lines (starting from the F2 generation) [Fig. 4c], purified total RNA from these lines, and performed RT-PCR assays to determine if any of these RNA binding proteins play a role in the splicing of these model alternative splicing events [Fig. 4d].
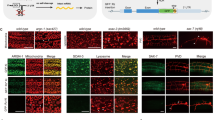
a Schematic of the two-color reporter system. Upon inclusion of the alternative exon (yellow box), the reading frame encoding mCherry is utilized, while the reading frame encoding GFP is not, indicated by the color change of the reading frame encoding GFP from deep green to light green. The exon skipping leads to a frameshift (+1) and translation of the GFP protein. Translated fluorescent proteins are uncoupled from upstream sequence through 2A peptides, and localized to the nucleus through two nuclear localization signals. Reporter-specific RT-PCR priming sites are also indicated in diagram. b Cocktail of four different splicing minigene reporters injected into worms to generate stable extrachromosomal array lines for the RNA collection and RT-PCR assays. c Stable extrachromosomal array lines generated from the injected mutants (RB1766): (i) bright field and fluorescent images of original mutant (RB1766). Images are representative of RB1766 from which 100% of animals show no fluorescent signals, which was selected for the injection. (ii) both GFP and mCherry signals observed in the neurons and intestine cells showing the transgenic progeny from injected mutants (RB1766) are successfully generated. Images are representative of at least 10 transgenic progeny obtained from the successful robotic injection of a cocktail of four plasmids into RB1766. d RT-PCR assays of the zoo-1 exon 9 reporter for selected six mutant strains. Images are representative of 5 RT-PCR assays of the zoo-1 exon 9 reporter for selected six mutant strains. Source data are provided as a Source Data file.
Several of the RNA-binding protein mutants are unhealthy, with reduced brood sizes potentially affecting the transformation efficiency. As such, at least 20 worms per mutant strain were injected to get enough transgenic F1 progeny allowing for transmitting stable transgenes arrays to the F2 generation, and in total more than 1000 worms were injected using the robotic system, further confirming the robustness of our microinjection system. We were able to establish transgenic animals stably inheriting reporter arrays for 48 of the 49 wild types and mutant strains injected. The one exception, lin-28(n719), was quite unhealthy and unable to lay eggs outside the body. Even if the transgenic F1 progeny were generated, they stayed inside the body of injected parent worms. Thus, its small brood size and failure to lay embryos made it very difficult to obtain stable extrachromosomal lines. We purified total RNA from all strains and screened for splicing pattern differences by semi-quantitative RT-PCR using primers specific for each reporter, comparing wild-type and mutant patterns [Supplementary Data 2]. In total, we performed over 250 RT-PCR assays in our screen.
After conducting densitometry [Supplementary Data 2] and a visual inspection of gel images, we selected six mutant strains of interest with detectable changes in splicing patterns compared to wild-type worms which are D1037.1(ok1746) I, rsp-2(ok639) II, tdp-1(ok803) II, cey-4(ok858) III, ptb-1(gk113) II, Y57G11C.36(gk5042) IV), respectively, for a replicate smaller scale screen (see “Methods”). In this second round, we also stage-synchronized all animals and collected populations at L4 stage to eliminate potential splicing differences obtained due to mixed population measurements from the primary screen. Intriguingly, we identified that deletion of tdp-1 led to a significant increase of exon skipping in our zoo-1 exon 9 reporters by 13.4% when compared to wild-type worms [Fig. 4d and Supplementary Data 3; mean percent spliced in (PSI) wild-type = 43.9\(\pm\)4%, mean PSI tdp-1 mutant = 30.5\(\pm\)5%, N = 5; t-test: p = 0.0023]. Thus, our screen has identified a putative regulator of a neuronally repressed alternative exon, which will warrant further study. More importantly, our results indicate that our robotic microinjection system can facilitate large-scale reporter screens in available knockout mutants.
Discussion
In this study, we have developed a robust robotic microinjection system with a high injection speed, high success rates, and high quality of worm injection required for high-throughput generation of transgenic C. elegans. While several robotic microinjection systems have been reported previously, the random orientation of worms, complex operation procedures, severe squeezing of loaded worms, and easy clogging and breakage of injection needle when inserted into the hydrogel, as shown in Supplementary Table 3, would prevent the precise and efficient microinjection of plasmid into the target body structure, further limiting their use in practical applications. Compared to manual microinjection experiments where operators must undergo a long period of training to become proficient and where it is difficult to perform injections for more than 2–3 h, it is very easy for unskilled operators to learn the usage of our robotic system after several trials and perform the worm injection with high success rate and excellent quality. Moreover, the robotic system minimizes the efforts, input, and fatigue by the user which allows for a fast injection speed while maintaining a consistent high-level performance. It has been demonstrated that the developed robotic system can be employed to realize single gonad arm injection of >250 worms per day when accommodating multiple strains and injection solutions. It should be noted that our robotic system can be easily adapted to the injection of different mutant strains of varying body sizes, showing great potential in a number of applications (e.g., genetic interaction profiling, genetic screens, and nucleic acid or small molecule delivery to the gonad) that require delivery of genetic or other solutions into mutants. In addition, the robotic system is also capable of providing secure immobilization of worms for the successful injection of the different genetic solutions, Furthermore, similar to the manual injection method, the robotic microinjection system manipulates worms in an open environment, allowing for the easy access of worms and preventing the frequent clogging of injection pipettes, which will make the present system easy to adopt by researchers from worm biology laboratories or core facilities providing microinjection services. Using our system, different types of genetic solutions, including plasmid and ribonucleoprotein complexes, have been successfully injected into the gonad of C. elegans to generate stably transmitting extrachromosomal arrays as well as genome-edited animals via CRISPR/Cas9 approaches.
Moreover, through a large-scale multiplexed plasmid reporter screen in knockout mutants, we discovered that the deletion of the tdp-1 gene leads to increased exon skipping in our zoo-1 exon 9 reporters, highlighting the speed and reliability of our platform in facilitating screens involving higher throughput microinjection. Alternative splicing patterns are known to be regulated in a combinatorial manner by RNA-binding proteins (for examples see refs. 44,45,46). As such, it is common for perturbations of individual RNA binding proteins to result in modest but significant effects on splicing patterns, such as what we have observed in tdp-1 mutant animals. Recently, we identified the conserved RNA binding protein CELF/UNC-75 as a repressor of zoo-1 alternative exon 9 splicing30. Our current results suggest that TDP-1 may be acting directly or indirectly to stimulate exon inclusion, counteracting the effects of UNC-75. Thus the balance of these RNA binding proteins, and likely others yet to be identified, in different tissues may provide an explanation for the predominant skipping of zoo-1 exon 9 in neurons and its preferential inclusion in other tissues30.
Due to the sophisticated structures inside the worm body, changing position of the distal gonad at different orientations, and the determined optimal vertical position of the mounted injection pipette, it is necessary to rotate the worm to an orientation in which the mounted injection pipette can approach the anterior/posterior distal gonad for material delivery. Our robotic system can overcome this important bottleneck in the microinjection procedure by achieving controlled rotation of animals in an open environment to position the distal gonad opposite the injection needle while providing secure immobilization of orientated worms for easy injection thereafter [Supplementary Table 4] which is impossible for other existing worm rotation methods18,19,20,47,48 and any existing worm injection platforms16,17,21. It should be noted that different from other rotation methods that require specific substrate designs47,49, our worm rotation method is simple, reliable, and independent of the substrate which means it can be easily adopted in many applications, especially those requiring a thin cover slip as the sample substrate for high-resolution imaging under high magnification. In addition, the combination of frictional force and hydrodynamic forces induces the rotational torque for contactless, precise, and stable rotation of a C. elegans, which allows for the worm rotation independent of the worm body size and curvature, and the level of anesthesia. Thus, the developed worm rotation method can be employed to realize stable rotation of different mutant strains with varying body sizes or harboring resistance to levamisole treatment. Stable worm rotation in a continuous fashion can be obtained by adjusting the operation parameters of the motorized stage and the pressure regulator. Moreover, a consistent gap between worm body and the microfluidic device during the rotation process is kept by the hydrodynamic forces caused by the laminar flow flowing out of microfluidic channels allowing for the clear observation of all anatomical regions of the animal, which provides accurate visual feedback for the worm orientation control. The open ends of microfluidic device enable the rotated worm to be smoothly and securely immobilized for versatile manipulations of worms including injection, biomechanics characterization, genetic screening, phenotyping, and microsurgery, to name just a few. Thus, the synergy of developed robotic system and microfluidic devices has opened a route to the accurate, high-efficiency, and intelligent microinjection of C. elegans and provided an opportunity to facilitate high-throughput transgenic studies of C. elegans.
Another major benefit of our developed robotic system is that it is compatible with all microscopes equipped with a motorized stage. In our robotic system, the microfluidic device was used for worm rotation and immobilization. As the simple microchannels of the microfluidic device have an array of open ends, any impurities which are brought into the channels during the injection can simply be ejected by applying a relatively small positive pressure. Thus, combined with its easy and cheap fabrication, the microfluidic device is highly reusable.
In basic biological mechanism studies, genetic and reporter library screening often requires large number of transgenic strains. Our robotic system will help alleviate the bottleneck of strain construction through its high speed and high injection success rate. Thus, researchers could envision performing screens involving microinjection on a larger scale, taking advantage of the growing toolbox of available transgenesis approaches in C. elegans50. Other large-scale studies like drug screening and toxicity can be easily realized by injecting candidate materials, especially water-insoluble materials, into specific organs using the developed system. Finally, in addition to C. elegans, this robotic system can be adapted for high-speed injection of Drosophila larvae or zebrafish larvae, another two important and popular small model organisms which have similar morphologies when comparable to C. elegans. To realize this goal, one would need to simply change the geometry (for example, height and width) of the microfluidic device while using the same basic features and function. Similarly, retuning parameters for larvae rotation is straightforward while all other components of the robotic system can remain the same. Taken together, this customizability affords our robotic system a wide range of applications. In addition, as machine learning technology continues to advance, bringing enhanced capabilities in learning complex patterns, fitting models to large and diverse datasets, and realizing accurate semantic segmentation and key-point detection tasks, it offers the opportunity to fully automate the microinjection of C. elegans in a reliable and consistent manner in the future.
Methods
Ethical statement
This research complies with all relevant ethical regulations. C. elegans does not fall under animal welfare regulations. Husbandry and housing were approved by local authorities under biosafety permit 527-C25-1 issued by the University of Toronto.
Microfluidic device fabrication
In this robotic system, a microfluidic device for stable worm rotation and secure immobilization was integrated as a greatly important part. Dimensions of the bifurcated channels inside microfluidic device are shown in Supplementary Fig. 9a. This microfluidic device was fabricated through multilayer soft lithography. First, SU-8 2075 photoresist (Microchem) with thickness of 25 μm was spin-coated onto a silicon wafer, and then patterned into the SU-8 mold of the top PDMS layer by standard photolithography, as shown in the Supplementary Fig. 9b-(i). This mold was further treated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane via chemical vapor deposition for one hour to render its surface hydrophobic. After that, a precursor mixture of PDMS base and cross-linker (Sylgard 184, Dow Corning) at the w/w ratio of 5:1 was poured onto the SU-8 mold and cured at 80 °C for 45 min [Supplementary Fig. 9b-(i)]. In the following, a precursor mixture of PDMS base and cross-linker (Sylgard 184, Dow Corning) at the w/w ratio of 10:1 was spin-coated onto a silicon wafer with the thickness of 14 μm [Supplementary Fig. 9b-(ii) and Supplementary Fig. 10] and cured at 80 °C for 45 min. Then, the top PDMS layer was peeled from the SU-8 molds, punched with open inlet holes, and bonded to the 14 μm-thick PDMS layer together to form the microfluidic device [Supplementary Fig. 9b-(iii)]. Later the microfluidic device consisting of the top PDMS layer and the bottom PDMS layer was peeled off from the silicon wafer [Supplementary Fig. 9b-(iv)]. To realize stable and reliable worm rotation, a smooth slope of 45° was cut at the side wall of the microfluidic device where the open ends of the microchannels are arranged, as shown in Supplementary Fig. 9c. Before cutting, the microfluidic device was securely clamped with assistance of bench vise, glass slides, and a 3D printed stage with a slope of 45° [Supplementary Fig. 9d]. Once the microfluidic device was securely clamped by the bench vise, a surgical scalpel was used to cut the microfluidic device along the slope of 3D printed stage and the edge of glass slide [Supplementary Fig. 9e]. After that, the microfluidic device was bonded to a transparency film (Inkjet Transparency Film, Staples) via a double-sided tape, thickness of which is 25 μm (8171CL, 3 M) [Supplementary Fig. 9b-(v), 9b-(vi)]. In this method, a transparency film was utilized as a supporting layer for the PDMS microfluidic device because it possesses a relatively rigid and flat surface, which prevented the bottom PDMS layer from adhering to the upper side of microchannels. Furthermore, the thickness of the transparency film is approximately 0.1 mm, thin enough to ensure that the bottom side of the microfluidic device at open ends can make contact with the substrate surface once it is lowered. Subsequently, a pre-cut cast acrylic sheet (PMMA) was bonded to the bottom side of transparency film using the double-sided tape, serving as a holder when it was mounted on the right micromanipulator (thickness: 1/16″, McMaster-Carr) [Supplementary Figs. 2g, 9b-(vii) and 9b-(viii)]. Finally, we found that fabricated microfluidic device can be tightly bonded to PMMA plate and reliably mounted on the right micromanipulator. The exploded view of whole microfluidic device was displayed in Supplementary Fig. 2g.
C. elegans strains, culture, and sample preparation
Both wild-type and transgenic worms [Supplementary Data 1 and Supplementary Table 5] were used in the rotation and microinjection experiments. Worm strains used for the regular/mixed plasmid DNA and CRISPR/Cas9 plasmid microinjection experiments were wild-type strains. The genetic strain for the RNP complex injection is JAC644 (Genotype: sup-46(csb40)[loxP + myo-2p::GFP::unc-54 3′UTR + rps-27p::neoR::unc-54 3′UTR + loxP]), in which the GFP signal is expressed in pharynx, as shown in Fig. 3d-(i). Synchronous populations of young adult nematodes were obtained for the injection using the following protocol. Briefly, three adult worms were first transferred to the nematode growth media (NGM) plate seeded with the bacterial food E coli OP50-1, and left on the plate for three days to collect a large population of adult worms from the first generation. Before loading the worms in the reservoir using a Pasteur pipette (9-inch, Fisher brand) for microinjection, adult worms were selectively picked up by a worm pick (made from a Pasteur pipette and a platinum wire) and suspended in the levamisole solution at a concentration of 0.6 mg mL−1 for 10–30 min to ensure that anesthetized animals body remained straight during the whole worm injection process. The levamisole solution of 0.6 mg mL−1 was prepared by mixing the levamisole solution in a concentration of 3 mg mL−1 and M9 buffer in a volume ratio of 1:4. To avoid anesthetized worms sticking to the surface of Pasteur pipette, Triton X-100 was added in the M9 buffer and the final concentration of Triton X-100 in M9 buffer is 0.01%. Once a batch of worms was injected, they were transferred to the NGM agar plates for maintenance. For the regular plasmid DNA injection, injected worms were cultured on NGM agar plates at 21 °C. After 2–4 days, transgenic F1 progeny can be observed using the fluorescent microscopy (Axio Zoom.V16, Zeiss) if the injection is successful. For the injection of CRISPR/Cas9 plasmid DNA, injected worms were cultured on NGM agar plates at 25 °C for one day followed by selection with the drug of G418, as shown in Supplementary Fig. 21. During the selection phase, we daily screened the plates to check the fluorescent pattern from the progeny. The recombinant animals which survived the G418 selection and possessed target fluorescent signal in a uniform fashion were then easily identified. The injection needle was obtained by pulling the borosilicate glass capillaries (outer diameter: 1.0 mm, inner diameter: 0.58 mm, 1B100F-4, World Precision Instrument) via the needle puller (P-97 micropipette puller, Sutter Instrument). Usually, the directly pulled injection needle has a closed tip. To make the tip open, the pulled injection needle is always pushed against a glass filament which would break the closed tip in a precise fashion. During the manual injection, an eyelash pick was used by the majority of experts and all proficient operators for worm handling which avoids the damage to worms. Fluorescent images of the transgenic worm were obtained by using inverted microscope (ZEISS Axio Observer, Zeiss).
Splicing reporter screens
For our screen, more than 20 young adult wild-type or mutant worms per strain (see Supplementary Data 1 for list of mutant strains tested) were injected as described above with a cocktail of four plasmids (Prgef-1::zoo-1 exon 9, Prgef-1::hlb-1 exon 12, Prgef-1::ampd-1 exon 15, and Pges-1::fhod-1 exon 9). All reporter plasmids were injected at a final concentration of 50 ng/μL. Three to four days after microinjection, 24 mCherry and GFP positive F1 progeny were picked for each strain using the fluorescent microscopy (ZEISS Axio Zoom.V16) and then F2 animals were inspected for inheritance of the mCherry and GFP marker to establish stable extrachromosomal array lines.
For the primary screen, one representative line was expanded for the wild-type and mutant strains, and then 12 transgenic young adults were placed on 4 plates and mixed-stage animal populations were collected 3–4 days later (upon inspection of population growth) and placed directly in 400 μL of Tri Reagent (Sigma Aldrich) and frozen at −80 °C. Total RNA was then purified as recommended by the manufacturer. RT-PCR assays were performed using the OneStep RT-PCR kit (Qiagen) as recommended by the manufacturer [Supplementary Data 4]. 50 ng of total RNA was used as input and between 30–35 cycles of amplification were used depending on the reporter and condition. All products were resolved on 2% agarose gels and imaged on a GelDoc (Bio-Rad) imaging system.
Densitometry was performed on background subtracted intensity values using ImageJ and a percent inclusion value was calculated as follows:
After inspecting densitometry measurements and visually inspecting gel images, a subset of mutants with detectable changes in splicing patterns compared to wild-type worms (D1037.1(ok1746) I, rsp-2(ok639) II, tdp-1(ok803) II, cey-4(ok858) III, ptb-1(gk113) II, Y57G11C.36(gk5042) IV) were selected for a replicate smaller scale experiment to re-test results from the primary screen. Microinjections were repeated and independent stable extrachromosomal array lines were obtained. This round, we also stage synchronized all animals and collected populations at L4 stage to eliminate potential splicing differences obtained due to mixed population measurements from the primary screen. Total RNA collection and RT-PCR assays were performed as described above, with the exception that the number of amplification cycles was reduced to 30 in order to minimize over-amplification of target complementary DNA (cDNA). Densitometry was once again performed as described above.
System control and image processing
We interfaced all components integrated into the robotic system to a computer workstation (Dell Precision 7820, 8-Core 2.1 GHz CPU) with a data acquisition card (Model 826, Sensoray). These components consist of an inverted microscope (Olympus IX83), a digital camera (scA640-74fc, Basler), two motorized micromanipulators (left micromanipulator: MP285; right micromanipulator: MX7600R), a pneumatic microinjector (IM-300, Narishige) mounted on the left micromanipulator, a microfluidic device mounted on the right micromanipulator, and a pressure regulator for providing a negative pressure to the microfluidic device inlet. The custom-made reservoir filled with M9 medium was placed on a motorized X-Y stage (ProScan-H117, Prior Scientific), and its movement was regulated via the motorized stage. The movement of pneumatic injector was controlled via the left micromanipulator, and the position of microfluidic device was precisely controlled via the right micromanipulator. The injection pressure provided by the pneumatic microinjector was set at 60 PSI. All components were controlled by a custom-designed C++ program. In addition, the C++ code also allowed for the image acquisition and image processing which provided visual feedback to the closed-loop control of injection pipette movement [Supplementary Fig. 7], enabled the variance computation in grey level among image pixels for autofocusing under 10× objective [Supplementary Fig. 5], and determined injection pipette home position based on motion history image algorithm [Supplementary Fig. 6].
Statistics and reproducibility
All the statistical analysis has been executed using Microsoft Excel (Microsoft 365), and the data of worm injection are visualized by using the Origin (OriginPro8.5.1, Originlab). For the validation of system performance, a minimum of ten worms were injected independently under different experimental conditions. Further, different plasmid DNA solutions were injected to support the efficiency of robotic injection system. All microinjection results presented in the figures depict the average of at least 10 independent experiments and their associated standard deviation. No statistical method was used to predetermine sample size. No data are excluded from the analyses. The adult worms of each strain were randomly selected for injection, and imaging of all the samples was randomly allocated. The investigator was not blinded to group allocation during the experiments. To study the effect of the volume of injected plasmid DNA solution on the generation of transgenic, we carried out six tests (50 pL, 100 pL, 200 pL, 300 pL, 400 pL, 500 pL). Single-factor ANOVA was performed to analyze difference among the average number of transgenic F1 progeny with different injection volumes. Two-sided t-test with equal or unequal variance which were first validated by the p value of F-test were conducted to compare the number of transgenic F1 progeny obtained from robotic injection and manual injections. Two-sided t-test with equal or unequal variance which were first validated by the p value of F-test were also performed to compare the number of transgenic F1 progeny obtained from robotic injection w/wo rotation process or by different operators.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All of the data in this work are presented in the main text and the Supplementary Information. Source data are provided with this paper.
Code availability
Code associated with the hardware manipulation and robotic microinjection is available at https://github.com/MBL-Group/Ro-Injection and on Zenodo51.
References
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat. Genet. 33, 228–237 (2003).
Shaye, D. D. & Greenwald, I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One 6, e20085 (2011).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. L. B Biol. Sci. 314, 1–340 (1986).
Stirman, J. N. et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat. Methods 8, 153–158 (2011).
Dong, X. et al. Toward a living soft microrobot through optogenetic locomotion control of Caenorhabditis elegans. Sci. Robot. 6, eabe3950 (2021).
Cho, Y. et al. Multimodal stimulation in a microfluidic device facilitates studies of interneurons in sensory integration in C. elegans. Small 16, 1–10 (2020).
Chew, Y. L. et al. An afferent neuropeptide system transmits mechanosensory signals triggering sensitization and arousal in C. áelegans. Neuron 99, 1233–1246 (2018).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Xu, T. et al. Descending pathway facilitates undulatory wave propagation in Caenorhabditis elegans through gap junctions. Proc. Natl Acad. Sci. 115, E4493–E4502 (2018).
Cuentas-Condori, A. et al. C. elegans neurons have functional dendritic spines. Elife 8, e47918 (2019).
Ji, N. et al. Corollary discharge promotes a sustained motor state in a neural circuit for navigation. Elife 10, e68848 (2021).
Tzur, Y. B. et al. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195, 1181–1185 (2013).
Norris, A. D., Kim, H.-M., Colaiácovo, M. P. & Calarco, J. A. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201, 449–458 (2015).
Gilleland, C. L., Falls, A. T., Noraky, J., Heiman, M. G. & Yanik, M. F. Computer-assisted transgenesis of Caenorhabditis elegans for deep phenotyping. Genetics 201, 39–46 (2015).
Dong, X., Song, P. & Liu, X. Automated robotic microinjection of the nematode worm Caenorhabditis elegans. IEEE Trans. Autom. Sci. Eng. 18, 850–859 (2020).
Pan, P. et al. On-chip rotation of Caenorhabditis elegans using microfluidic vortices. Adv. Mater. Technol. 6, 2000575 (2021).
Ahmed, D. et al. Rotational manipulation of single cells and organisms using acoustic waves. Nat. Commun. 7, 11085 (2016).
Ozcelik, A. et al. Acoustofluidic rotational manipulation of cells and organisms using oscillating solid structures. Small 12, 5120–5125 (2016).
Zhao, X. et al. Microfluidic chip-based C. elegans microinjection system for investigating cell–cell communication in vivo. Biosens. Bioelectron. 50, 28–34 (2013).
Duffy, D. C., McDonald, J. C., Schueller, O. J. A. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly (dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Quake, S. R. & Scherer, A. From micro-to nanofabrication with soft materials. Science 290, 1536–1540 (2000).
Altun, Z. F., Hall, D. H. Introduction, in WormAtlas. https://doi.org/10.3908/wormatlas.1.1 (2009).
Wood, W. B., Bergmann, D. & Florance, A. Maternal effect of low temperature on handedness determination in C. elegans embryos. Dev. Genet. 19, 222–230 (1996).
Yoshida, N. & Perry, A. C. F. Piezo-actuated mouse intracytoplasmic sperm injection (ICSI). Nat. Protoc. 2, 296–304 (2007).
Dai, C. et al. Design and control of a piezo drill for robotic piezo-driven cell penetration. IEEE Robot. Autom. Lett. 5, 339–345 (2019).
Huang, H., Mills, J. K., Lu, C. & Sun, D. A universal piezo-driven ultrasonic cell microinjection system. Biomed. Microdevices 13, 743–752 (2011).
Norris, A. D., Gracida, X. & Calarco, J. A. CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. Elife 6, e28129 (2017).
Koterniak, B. et al. Global regulatory features of alternative splicing across tissues and within the nervous system of C. elegans. Genome Res. 30, 1766–1780 (2020).
Sellegounder, D. et al. Neuronal GPCR NPR-8 regulates C. elegans defense against pathogen infection. Sci. Adv. 5, eaaw4717 (2019).
McDiarmid, T. A. et al. Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc. Natl Acad. Sci. 117, 656–667 (2020).
Au, V. et al. CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditis elegans. G3 Genes Genomes Genet. 9, 135–144 (2019).
Berkowitz, L. A., Knight, A. L., Caldwell, G. A. & Caldwell, K. A. Generation of stable transgenic C. elegans using microinjection. J. Vis. Exp. JoVE, (2008).
Evans, T. Transformation and microinjection. WormBook 1–11 https://doi.org/10.1895/wormbook.1.108.1 (2006).
Dickinson, D. J., Ward, J. D., Reiner, D. J. & Goldstein, B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013).
Friedland, A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743 (2013).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Sioson, V. A., Kim, M. & Joo, J. Challenges in delivery systems for CRISPR-based genome editing and opportunities of nanomedicine. Biomed. Eng. Lett. 11, 217–233 (2021).
Prior, H., Jawad, A. K., MacConnachie, L. & Beg, A. A. Highly efficient, rapid and Co-CRISPR-independent genome editing in Caenorhabditis elegans. G3 Genes. Genomes Genet. 7, 3693–3698 (2017).
Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017).
WANG, Y. et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 3, 152–158 (2015).
Ule, J. & Blencowe, B. J. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell 76, 329–345 (2019).
Norris, A. D. et al. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 54, 946–959 (2014).
Wang, E. T. et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 25, 858–871 (2015).
Solana, J. et al. Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. Elife 5, e16797 (2016).
Zhang, J. et al. Surface acoustic waves enable rotational manipulation of Caenorhabditis elegans. Lab Chip 19, 984–992 (2019).
Ardeshiri, R., Mulcahy, B., Zhen, M. & Rezai, P. A hybrid microfluidic device for on-demand orientation and multidirectional imaging of C. elegans organs and neurons. Biomicrofluidics 10, 064111 (2016).
Chen, C. et al. Acoustofluidic rotational tweezing enables high-speed contactless morphological phenotyping of zebrafish larvae. Nat. Commun. 12, 1118 (2021).
Nance, J. & Frøkjær-Jensen, C. The Caenorhabditis elegans transgenic toolbox. Genetics 212, 959–990 (2019).
Pan. P., et al, Robotic microinjection enables large-scale transgenic studies of Caenorhabditis elegans: v0.0.1. Zenodo, https://doi.org/10.5281/zenodo.13624036 (2024).
Acknowledgements
The authors acknowledge financial support provided by the Natural Sciences and Engineering Research Council of Canada RGPIN-2017-06374 (X.L.), the Natural Sciences and Engineering Research Council of Canada RGPIN-2022-05039 (X.L.), the Canada Foundation for Innovation JELF-38428 (X.L.), and the Canadian Institutes of Health Research CIHR- 389077 (J.A.C.).
Author information
Authors and Affiliations
Contributions
Conceptualization and design: P.P., J.C., X.L. System setup: P.P., P.Z., W.S., R.Z., S.W., A.L, X.L. C. elegans maintenance and plasmid DNA preparation: P.P., M.Z., S.P., S.B., P.P.P., C.L., C.M., K.P., Robotic injection: P.P., C.L., P.X., Z.Z., R.L., Z.Q. Manual injection: M.Z., P.P.P., C.L., C.M., W.H., H.T., K.M., B.Y., J.A.C. Data collection and analysis: P.P., P.Z., M.Z., J.A.C., X.L. Writing: P.P., P.Z., W.B.D., M.Z., A.L.S., J.A.C., X.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Zihao Li and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, P., Zoberman, M., Zhang, P. et al. Robotic microinjection enables large-scale transgenic studies of Caenorhabditis elegans. Nat Commun 15, 8848 (2024). https://doi.org/10.1038/s41467-024-53108-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53108-5