Abstract
Bone Morphogenetic Protein (BMP) signaling is essential for craniofacial development, though little is known about the mechanisms that govern BMP secretion. We show that depolarization induces calcium-dependent BMP4 release from mouse embryonic palate mesenchyme. We show endogenous transient changes in intracellular calcium occur in cranial neural crest cells, the cells from which embryonic palate mesenchyme derives. Waves of transient changes in intracellular calcium suggest that these cells are electrically coupled and may temporally coordinate BMP release. These transient changes in intracellular calcium persist in palate mesenchyme cells from embryonic day 9.5 to 13.5 mice. Disruption of a potassium channel called Kcnj2 significantly decreases the amplitude of calcium transients and the ability of cells to secrete BMP. Kcnj2 knockout mice have cleft palate and reduced BMP signaling. Our data suggest that temporal control of developmental cues is regulated by ion channels, depolarization, and intracellular calcium for mammalian craniofacial morphogenesis.
Similar content being viewed by others
Introduction
Genetic syndromes demonstrate that ion channels contribute to human facial morphogenesis. Syndromes caused by disruption or activation of ion channel function are termed channelopathies. An analysis of these disorders demonstrates abnormal craniofacial development is associated with mutations that disrupt calcium channels (e.g., CACNA1C, Timothy syndrome)1,2,3, potassium channels (e.g., KCNK9-Birk-Barel syndrome; KCNJ2-Andersen-Tawil syndrome, GIRK2-Keppen-Ludinski syndrome)4,5,6,7,8,9,10,11,12,13,14, and sodium channels (e.g., NALCN-Infantile hypotonia with psychomotor retardation and characteristic faces (IHPRF))15. Furthermore, intrauterine exposure to teratogens that impact ion channel function, such as anti-epileptic drugs, heat, nicotine, and cannabinoids, is associated with increased risk for craniofacial defects15,16,17,18,19,20,21,22,23,24. Similarly, genetic inhibition of ion channels can cause morphological abnormalities in several species of animals25,26,27,28,29,30,31,32. Ion channels work together to establish the electrical properties, including the membrane potential of each cell. The diversity of ion channels that are important for craniofacial development suggests that cellular electrical properties impact morphogenesis. However, the molecular mechanisms by which membrane potential contributes to craniofacial morphogenesis remain unclear.
One of several possibilities is that ion channels impact the complex communication patterns between cells during facial morphogenesis. Proper formation of the face requires cranial neural crest (CNC) cells to migrate from the neural tube to populate the frontal nasal process and pharyngeal arches33,34. CNC cells form the craniofacial bone and cartilage, cranial neurons and glia, odontoblasts, and melanocytes and require precise spatiotemporal control to produce the proper cell types35. This is accomplished through signaling networks of morphogens, such as Bone morphogenetic protein (BMP), Notch, and Sonic hedgehog (Shh)36,37,38. An impressive amount of research has revealed the underpinnings of these molecular signaling cascades. For example, BMP is an essential molecular signal for craniofacial development. Upon BMP ligand binding, receptors phosphorylate Smads which can then enter the nucleus to induce target gene expression39,40,41,42,43. BMP signaling also activates extracellular signal-regulated kinases (ERKs) for osteoblast differentiation44. Oscillatory, or pulsatile ligand exposure is key for BMP to communicate efficiently via receptor engagement and Smad activation45. Osteoblast development during regeneration relies on waves of oscillatory ERK activation in zebrafish46, suggesting that temporal regulation is important for both modes of BMP signaling. However, we know very little about how cells control the timing of molecular signals that mediate BMP release. If the temporal signaling pattern matters for the transcriptional output, how do cells control ligand release? One possibility is that ion channels control the pulsatile cellular release of ligands.
The established roles of ion channels in traditionally excitable cell types, such as neurons, may lend insight into the role of membrane potential in craniofacial development. In neurons, ion channels coordinate the precise release of vesicles containing molecular signals, called neurotransmitters, to orchestrate complex communication between cells47. This is accomplished when the cell’s membrane potential rapidly changes due to the influx of sodium and calcium, and the efflux of potassium ions controlled by the opening and closing of ion channels. Thus, an electrical signal coordinates the delivery of a chemical signal. Recent evidence suggests that ion channels may perform a similar function to control timing and release of developmental cues in cell types not classically thought of as excitable. Inwardly rectifying potassium channel KCNJ2 (Kir2.1) loss of function in humans, mice, and flies cause defects that are remarkably similar to those arising from a loss of BMP signaling26,28. In Drosophila, a homolog of Kcnj2 called Irk2 is required for the BMP homolog Decapentaplegic (Dpp)-mediated patterning of the wing28. Loss of Irk2 conduction disrupts pulsatile Dpp release in the wing primordium (wing disc)27. Further, Dpp release can be induced by depolarization27. In cell culture, pulsatile presentation of BMP ligands produces a greater transcriptional response than constant exposure45, suggesting an intimate link between coordinated timing of release and downstream patterning. This requirement for Kcnj2 homologs in BMP signaling is conserved, as Kcnj2 is important for efficient BMP signaling in mice26. Kcnj2 and BMP are required in cranial neural crest cells for craniofacial patterning, including palate development26,48. While BMP signaling is significantly reduced in mouse embryonic Kcnj2ko/ko palate cells, BMP ligands, receptors, and Smads are expressed at least at wild-type levels in the same cells26. These data suggest the enticing possibility that membrane potential controls BMP release with a similar mechanism to neuronal neurotransmitter release. Ion channels regulate membrane potential, which controls the calcium-dependent release of vesicles containing developmental signals.
Here, we investigate the hypothesis that ion channel-mediated membrane potential controls BMP release from palatal mesenchyme cells. We introduce a novel tool to visualize BMP4 release from cells from the developing mouse palatal mesenchyme. We show that depolarization of mouse embryonic palatal mesenchyme cells induces vesicular fusion of BMP-containing vesicles and this process is calcium-dependent. We show that depolarization stimulates BMP4 release from these cells. We demonstrate that CNC cells and embryonic palate mesenchyme cells exhibit endogenous transient changes in intracellular calcium that can propagate through waves between cells. Calcium transients are dependent on Kcnj2 and loss of Kcnj2 reduces BMP secretion. Together our results suggest ion channels can act upstream of BMP signaling in the developing mouse palate.
Results
Membrane potential regulates BMP release
Mutations in Kcnj2 cause cleft palate and other congenital defects and reduce BMP signaling in multiple organisms9,25,26,27,28,43. Because Kcnj2 regulates resting membrane potential, we use Kcnj2ko/ko mice as a tool to determine how membrane potential contributes to BMP signaling. While BMP signaling in developing palate cells of Kcnj2ko/ko mice is reduced, expression of BMP ligands, receptors, and smads is not reduced compared to wild-type26, so we hypothesized that Kcnj2 regulates BMP ligand release. We used cultured Kcnj2ko/ko and wild-type mouse embryonic fibroblasts (MEFs) to test this hypothesis because like palate cells, Kcnj2ko/ko MEFs express BMP ligands at least at wild-type levels, but MEFs are easily cultured and proliferate to confluency (Fig. 1A, B). We incubated WT mouse embryonic fibroblasts in conditioned media from WT, Kcnj2KO/+, or Kcnj2KO/KO cultured mouse embryonic fibroblasts and measured phosphorylated Smad 1/5 at serine residues 463/465 in the exposed WT MEFs (Fig. 1A). Smad 1/5 phosphorylation at serine residues 463/465 is a specific readout of BMP signaling. Smad 1/5 phosphorylation was significantly reduced in WT MEFs incubated with Kcnj2KO/KO conditioned media compared to cells treated with WT-conditioned media (Fig. 1B, C). There was a trend towards reduced phosphorylation of Smad 1/5 in WT cells treated with Kcnj2KO/+ conditioned media. Because BMP ligand expression was increased in Kcnj2KO/KO cells, but conditioned media contained less BMP for downstream signaling, we reasoned that perhaps membrane potential regulates BMP ligand release from palate mesenchyme cells as membrane potential regulates BMP release in neurons49. We needed a method to measure BMP release from palate cells to test this hypothesis.
A A diagram shows the experimental design. MEFs were isolated from WT, Kcnj2KO/+, and Kcnj2KO/KO E14 pups. RNA was collected from MEFs to determine the expression of BMP ligands by quantitative RT-PCR. Media was conditioned for 8 h with MEFs from E14.5 WT, Kcnj2KO/+, or Kcnj2KO/KO. Conditioned media was placed on WT MEFs for 24 h. Protein was isolated from treated WT MEFs for western blot analysis. B Quantitative RT-PCR shows that Kcnj2KO/KO MEFs express significantly more BMP6 than WT MEFs (P = 0.004 by two-tailed unpaired t test, N = 3 MEFs from independent WT embryos and 2 Kcnj2KO/KO). Error bars represent the standard deviation. C A representative western blot shows anti-rabbit P-Smad 1/5 and anti-GAPDH (loading control) from WT lysates incubated in conditioned media from MEFs from three different embryos of each genotype. Full images of both western blots are provided in the Source Data file. D A graph shows quantification of relative fluorescence of P-Smad 1/5/8 from western blots (P values attained via two-tailed unpaired t test between WT and Kcnj2KO/KO or Kcnj2KO/+ groups), N = 6 WT embryos, 5 Kcnj2KO/+, and 6 Kcnj2KO/KO. Source data are provided in the Source Data file.
Development of BMP4 release reporter
To examine the release of BMP4 from within BMP-producing mammalian cells, super ecliptic pHluorin (SEP), a pH-sensitive GFP variant, was inserted into the linker domain of BMP4 (Fig. 2A, see “Methods”), similar to the imaging tool used previously in D. melanogaster27. The fluorescence of SEP is quenched in acidic conditions, such as within a vesicle, and fluoresces in neutral conditions, such as when released into the extracellular environment. To test the function of this BMP4-SEP reporter, we utilized an immortalized mouse embryonic palatal mesenchyme (iMEPM) cell line44. iMEPM cells are immortalized cells with an increased proliferative capability but maintain palatal mesenchyme characteristics such as morphology, transcriptional landscapes, and migratory capability44. These cells are representative of the E13.5 mouse palate at an important time for signaling in the developing palate. iMEPMs can be used to investigate BMP signaling in mammals. To test the function of the reporter, iMEPM cells expressing BMP4-SEP were cultured for 24 h. Ammonium chloride (5 mM) was added to the cells during live imaging to neutralize the cellular pH without depolarizing them (Fig. 2B), and the resulting changes in SEP fluorescence were recorded (Fig. 2C)45. Upon neutralization, the iMEPM cells transfected with the BMP4-SEP construct had a robust increase in fluorescence, and this increase was significantly higher (P = 0.0019) than that of our empty vector control plasmid (pCIG-GFP) (Fig. 2C). These results are consistent with our expectations of the reporter construct and thus confirm the validity of the tool.
A Diagram of BMP4-SEP fusion protein. A pH-sensitive GFP variant, super ecliptic pHluorin (SEP), was inserted into the linker domain of BMP4 between Asn307 and Cys308 and cloned into a plasmid under control of a CMV promoter. B Diagram of iMEPM neutralization by ammonium chloride (NH4Cl). The acidic environment of a vesicle (red, pH ~5.5) restricts the fluorescence of SEP. When NH4Cl is applied to a cell, the environment becomes neutralized (pH = 7.4), unquenching SEP and increasing fluorescence visualization. C A violin plot shows a significant increase in fluorescence amplitude between iMEPM cells expressing BMP4-SEP compared to a pCIG-GFP control after neutralization by the addition of 5 mM NH4Cl (**P = 0.003 by unpaired two-tailed t test). D Diagram of BMP4-SEP release in response to cellular depolarization induced by the addition of KCl. E ViolinPlot showing quantification of SEP fluorescence in iMEPM cells transfected with pcDNA-SEP (blue), TfR-SEP (pink) or BMP4-SEP (green). Changes in fluorescence amplitude in regions of interest (ROIs) taken in live imaging videos (see methods) were averaged and compared. BMP4-SEP and TfR-SEP transfected cells had significantly higher changes in fluorescence than pcDNA-SEP (vs BMP4-SEP P = 0.017; vs TfR-SEP P = 0.0003) but were not found to be different from one another (P = 0.24). F–H Representative images (from seven plates of cells, two separate trials) show pcDNA-SEP, TFR-SEP, and BMP4-SEP fluorescence in iMEPM cells pre- and post-depolarization by 50 mM KCl. Red boxes denote the location of the magnified blue insets within each respective image. F’–H’ Representative fluorescence traces from iMEPM cells transfected with pcDNA-SEP, TFR-SEP, or BMP4-SEP construct. The addition of 50 mM KCl is denoted with the red arrow (**P < 0.05 ns=not significant by two-way ANOVA). Source data are provided as a Source Data file.
Depolarization induces vesicular release in iMEPMs
To test the hypothesis that palate mesenchyme cells are capable of depolarization-induced vesicular fusion for BMP release, we live imaged iMEPMs expressing a canonical marker of vesicular fusion, transferrin receptor-super ecliptic pHluorin (TfR-SEP)46,47. We compared iMEPMs expressing TfR-SEP and SEP alone as a control, during induced depolarization events (Fig. 2D). Fluorescence is dispersed throughout the cytoplasm of iMEPM cells that express SEP alone (Fig. 2F and Supplementary Movie 1). Upon depolarization, induced by the addition of 50 mM potassium chloride, we did not see stark increases in fluorescence in cells expressing SEP alone (Fig. 2F, F’ and Supplementary Movie 1). In contrast, depolarization of TfR-SEP expressing cells significantly increased punctate fluorescence intensity (Fig. 2G, G’ and Supplementary Movie 2). This punctate increase in SEP fluorescence was significantly greater than iMEPM cells expressing SEP in the cytoplasm (176.6 ± 19.60 vs. 407.4 ± 54.71 AU, P = 0.0034) (Fig. 2E). These data suggest that mouse palatal mesenchyme cells contain the machinery to respond to depolarization with vesicle fusion.
Depolarization induces BMP4 release from iMEPMs
To determine if depolarization can induce BMP4 release, we depolarized BMP4-SEP expressing iMEPM cells during live imaging. Depolarization induced a clear increase in punctate BMP4-SEP fluorescence (Fig. 2H, H’ and Supplementary Movie 3), supporting the model that depolarization can induce BMP4 release. Upon depolarization, BMP4-SEP fluorescence intensity increased significantly more than SEP in the cytoplasm in iMEPM control cells (176.6 ± 19.60 vs. 323.4 ± 46.42 AU, P = 0.0016) (Fig. 2E). Interestingly, the amplitudes of fluorescence change for BMP4-SEP were not significantly different from those observed using the canonical TfR-SEP exocytosis reporter. Both BMP4-SEP and TfR-SEP fluorescence appeared as punctate on the surface of iMEPMs after depolarization (Fig. 2E, G, H’). These results suggest that depolarization induces BMP4 release from iMEPM cells.
Depolarization increases BMP4 concentrations in iMEPM-conditioned media
An increase in BMP4-SEP fluorescence upon depolarization supports the hypothesis that depolarization can induce BMP release from iMEPM cells. To quantify and measure BMP release, we used a BMP ELISA. BMP4-SEP was transfected into iMEPM cells and conditioned media was collected before and after depolarization (Fig. 3A). Conditioned media collected after depolarization had BMP4 concentrations that were significantly greater than before depolarization (0.72 ± 0.08 vs. 0.95 ± 0.09 pg/mL, P = 0.0003, N = 20 plates of cells) (Fig. 3B). The increase in BMP4 concentration in media following depolarization suggests that depolarization induces BMP4 release from iMEPM cells. These data suggest that ion channels that regulate the membrane potential of palate mesenchyme cells also control BMP release. We know that depolarization causes transient increases in cytoplasmic calcium to induce fusion of vesicles and mediate ligand release in excitable cells like neurons. We next asked if depolarization increases cytoplasmic calcium in the palatal mesenchyme.
A A schematic created in BioRender107 shows the method used to quantify the amount of BMP4 in conditioned media before and after depolarization of BMP4-transfected iMEPM cells. iMEPM cells were cultured for 24 h before conditioned media was collected and frozen. Cells were depolarized with KCl solution. Immediately following depolarization, conditioned media was collected and frozen. An ELISA was conducted with paired conditioned media samples collected before and after depolarization. B A paired box-and-whisker plot shows a significant increase in the BMP4 concentration after depolarization by KCl (***P = 0.0003 by two-tailed paired t test, n = 20 plates of cells). Error bars represent minimum and maximum BMP concentration, horizontal line represents the median BMP concentration values, and the bounds of the box represent the 25th and 75th percentile. Source data are provided as a Source Data file.
Depolarization induces transient increases in cytoplasmic calcium in primary palatal mesenchyme
We expressed the fluorescent calcium reporter GCaMP6s in the palatal mesenchyme with Wnt1Cre. We dissected, dissociated, and cultured E13.5 palatal cells overnight. We imaged fluorescence over time during depolarization with KCl (50 mM). Depolarization significantly increased GCaMP6s fluorescence (n = 3 palates with 5–15 dissociated cells measured per palate, Fig. 4A–C and Supplementary Movie 4). Importantly, KCl-induced depolarization did not decrease a cell’s ability to recover and exhibit additional calcium transients (Fig. 4C). We imaged GCaMP expressing palate mesenchyme cells without depolarization to determine if cells undergo endogenous calcium events. Endogenous calcium events were observed in multiple fields of view in three independent cultured palates. The amplitude of KCl-induced transient increases in GCaMP fluorescence were significantly greater than endogenous events (Fig. 4D, unpaired t test). We observed transient increases in GCaMP fluorescence with depolarization in the same plate of cells multiple times after cells were reacclimated in culture media for 1 h between experiments. We conclude that depolarization increases cytoplasmic calcium in E13.5 mouse palatal mesenchyme.
A Representative images show that depolarization by the addition of KCl at 20 s increases the fluorescence of GCaMP6 expressed in dissociated primary cultured E13.5 palate mesenchymal cells (blue stars), and cells have subsequent calcium events following depolarization (red arrows). Replicates=3 plates depolarized with KCl, transient changes in GCaMP fluorescence measured in 36 cells. B Representative fluorescence profile of one cell over time out of 36 cells with transient increases in GCaMP fluorescence. C Representative profile of fluorescence over time for a cell that undergoes a calcium transient with depolarization at 20 s followed by two endogenous transients. D Depolarization induces significant increases in fluorescence compared to background changes in fluorescence N = 3 plates of primary culture MEPMs imaged before (control) and after depolarized with KCl (experimental), violin plot represents 38 ROIs from the three plates before depolarization and 36 ROIs measured with depolarization (****P = 5.7 × 10−12 by two-tailed unpaired t test). E Mean fluorescence/F0 traces of BMP4-SEP release averaged between cells treated with or without BAPTA-AM. Yellow represents DMSO controls (n = 6 from separate plates), and blue represents BAPTA-AM treated cells (n = 10 cells from separate plates). The red arrow denotes the addition of isosmotic KCl Tyrode’s media to induce depolarization. SEM is shown with shaded areas. F A box-and-whisker plot quantifying change in BMP4-SEP fluorescence amplitude over F0 between DMSO controls and BAPTA-AM treated iMEPM cells. The error bars represent minimum and maximum fluorescence values, and the center line represents the median value (*P = 0.0002 by unpaired two-tailed t test). Source data are provided as a Source Data file.
Depolarization does not affect cell viability or future calcium activity
We measured cell viability with crystal violet assays after depolarization with potassium chloride. Cell viability was measured with crystal violet staining and showed intact, healthy iMEPMs after treatment with 50 mM, 100 mM, and 150 mM KCl (Fig. S1). Next, we asked if depolarization by our method disrupted native electrical activity of primary culture MEPMs. We show that depolarized cells have subsequent calcium release events (Fig. 4C). Finally, after depolarization, we incubated cells in culture media overnight and imaged again the next day to confirm cells remained intact and undergo calcium transients.
BMP4 release from iMEPMs is calcium-dependent
To determine if depolarization-induced BMP release is dependent on cytoplasmic calcium, we repeated the BMP4-SEP experiments under isosmotic conditions with and without a cell-permeant calcium chelator called BAPTA-AM (Fig. 4E, F). Cells were incubated in 100uM BAPTA-AM for one hour to chelate intracellular calcium50. None of the BAPTA-AM treated IMEPM cells had large increases in BMP4-SEP fluorescence, indicating that calcium chelation inhibits BMP4-SEP release. Additionally, we found that calcium chelation reduced the number of cells in which we observed even a small increase in BMP4-SEP fluorescence upon depolarization from six out of twelve control cells (50%) to two out of ten (20%) treated cells. In the BAPTA-AM loaded cells, changes in the amplitude of BMP4-SEP fluorescence upon depolarization were significantly reduced from cells without BAPTA-AM treatment (19,448 ± 6079 vs. 4092 ± 2141 AU, P = 0.039) (Fig. 4E, F). Together, these data demonstrate that cytoplasmic calcium is necessary for depolarization-induced BMP release from palatal mesenchyme cells.
Palate mesenchyme undergoes endogenous transient changes in intracellular calcium
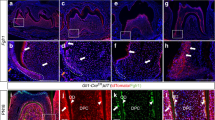
For coordination of morphogenesis, cells need to send and receive precise signals. Neurons achieve precise temporal control of molecular signals using depolarization to control transient changes in intracellular calcium to drive vesicular fusion. To investigate whether transient changes in intracellular calcium occur at a time of active signaling for palatogenesis, we cultured dissociated E13.5 primary mouse embryonic palate (MEP) cells that express the calcium sensor, GCaMP6s, driven by Wnt1Cre and imaged fluorescence over several minutes. These experiments revealed calcium transients (Fig. 5A and Supplementary Movie 5) and demonstrated that calcium events in the palate can be periodic (Fig. 5B (yellow trace)). An average of 3.24 ± 0.16 events occurred in cells over the imaging period (384 s) (Fig. 5E). This number varied, however, with some cells experiencing only one event and others undergoing twelve events in that time. Calcium transients had an average fold change in amplitude (ΔFluorescence/F0) of 3.47 ± 0.091 ΔF/F0 (Fig. 5D). Calcium transients vary in intensity with some cells exhibiting high-amplitude changes in GCaMP fluorescence while others show lower amplitude changes (Figs. 5A and S2). In cells experiencing repeated periodic events, average interevent period was 63.22 ± 2.50 s (Fig. 5F).
A Wnt1CRE; GCaMP6s expressing E13.5 MEP primary culture illustrating strong (red arrow) and weak (blue arrow) calcium activity in two cells. Calcium transients measured in 197 Wnt1CRE; GCaMP6s cells. B Representative GCaMP6s fluorescence traces of Wnt1CRE; GCaMP6s cells (yellow), Wnt1CRE; GCaMP6s; Kcnj2KO/+ cells (green), and Wnt1CRE; GCaMP6s Kcnj2KO/KO cells (orange) E13.5 MEPM cells. Calcium transients measured in 197 WT (Wnt1CRE; GCaMP6s) cells, 158 Kcnj2KO/+ cells, and 102 Kcnj2KO/KO Wnt1CRE; GCaMP6s cells. C Quantification of GCaMP6s initial fluorescence showing a significant reduction in Wnt1CRE; GCaMP6s; Kcnj2KO/+ (P = 1.37 × 10−18), and Wnt1CRE; GCaMP6s Kcnj2KO/KO (P = 1.044 × 10−13) compared to controls that have two wild-type Kcnj2 alleles. D Graph shows a significant reduction in amplitude of GCaMP fluorescence changes in Wnt1CRE; GCaMP6s; Kcnj2KO/+ (P = 9.5 × 10−21), and Wnt1CRE; GCaMP6s Kcnj2KO/KO (P = 5.3 × 10−25)compared to controls that have two wild-type Kcnj2 alleles. E, F Quantification of GCaMP event period and a number of events show no change between genotypes. (****P < 0.00001, ns=not significant by one-way ANOVA). Source data are provided as a Source Data file.
Kcnj2 mediates endogenous calcium transients in mouse embryonic palate primary cell cultures
To determine if a channelopathy-associated ion channel (Kcnj2) regulates palatal calcium transients, we quantified calcium transients in primary MEPM cultures from Wnt1Cre; GCaMP6s; Kcnj2KO/+ and Wnt1Cre; GCaMP6s; Kcnj2KO/KO mice compared to cells from Wnt1Cre; GCaMP6s control mice. When compared to Wnt1Cre; GCaMP6s littermates, loss of one or both copies of Kcnj2 reduced the initial GCaMP6s fluorescence within the cells, defined as F0 (Table 1, Fig. 5C, and Supplementary Movies 5–7). The mean amplitude of the calcium transients was reduced in Kcnj2KO/+ and Kcnj2KO/KO palate cells (Table 1 and Fig. 5B, D). However, the interevent period was not perturbed in either Kcnj2KO/+ or Kcnj2KO/KO palate cells (Table 1 and Fig. 5F). Interestingly, the mean number of events per cell was not significantly different between conditions in either Kcnj2KO/+ or Kcnj2KO/KO (Table 1 and Fig. 5E).
Cranial neural crest cells undergo calcium transients that are controlled by Kcnj2
Because mutations in Kcnj2 cause an array of craniofacial phenotypes in addition to cleft palate, we hypothesized that endogenous transient changes in intracellular calcium may be common to cranial neural crest (CNC) cells, the precursors of several craniofacial skeletal structures. To determine if transient changes in intracellular calcium occur in CNC cells, we cultured and live imaged E9.5 Wnt1Cre, GCaMP6s primary explants containing CNC cells. Live imaging of CNC cells that express GCaMP6s revealed periodic calcium transients in single cells with an average amplitude of 1.80 ± 0.14 F/F0 (n = 89 events) (Fig. 6A, C, D). Interestingly, we observed instances where groups of neighboring cells display synchronized increases in GCaMP6s fluorescence and in close succession. We call synchronized events calcium waves (Fig. 6B, C). To investigate if loss of Kcnj2 perturbed CNC cells calcium transients, we compared GCaMP6s fluorescence in Wnt1Cre; GCaMP6s control mice to Wnt1Cre; GCaMP6s; Kcnj2KO/+ and Wnt1Cre; GCaMP6s; Kcnj2KO/KO mice. We found that the loss of both copies of Kcnj2 significantly reduced the amplitude of GCaMP6s fluorescence, Fig. 6D, while the loss of only one copy of Kcnj2 did not significantly decrease fluorescence amplitude (Fig. 6D). Amplitudes of Kcnj2KO/KO CNC cells calcium transients were significantly lower than Kcnj2KO/+ transients (P = 0.04). We did not see a significant difference in F0 between any of the conditions (Fig. 6E). To determine if iMEPM cells undergo transient changes in intracellular calcium, we transfected iMEPM cells with a plasmid expressing GCaMP6s. We observed calcium events indicated by changes in GCaMP6s fluorescence in iMEPM cells (Fig. S3). These results support the hypothesis that CNC cells and their derivative cells undergo transient changes in intracellular calcium that may result in the release of BMP4.
A Wnt1CRE; GCaMP6s expressing E9.5 CNC cells explants showing calcium transients (red arrows) representative cells out of 198 E9.5 Wnt1CRE; GCaMP6s cells measured. B Wnt1CRE; GCaMP6s expressing E9.5 CNC cells explants showing a wave propagation event (centered on red asterisk). Synchrony analyzed in Fig. 7. C An example of mean GCaMP fluorescence plot for an E9.5 CNC cells cell. D Quantification of GCaMP6s amplitude showing a significant reduction in Wnt1CRE; GCaMP6s; Kcnj2KO/KO compared to controls (1.80 ± 0.14 vs. 0.91 ± 0.10 F/F0 for Kcnj2KO/KO, P = 0.019) while Kcnj2KO/+ did not significantly reduce amplitude (1.80 ± 0.14 vs. 1.60 ± 0.11 F/F0 for Kcnj2KO/+, P = 0.93. E Quantification of GCaMP initial fluorescence shows no change from controls. (*P < 0.0332, ns=not significant by two-way ANOVA). Source data are provided as a Source Data file.
Cranial neural crest-derived cells are electrically coupled
We observed that several cranial neural crest cells undergo transient changes in intracellular calcium together, suggesting that they are electrically coupled. To quantify the synchronization of these transient changes in intracellular calcium, we measured calcium activity throughout the area of the explant. We determined the proportion of the area that was synchronously active. We found that 32% of the total area exhibited synchronized calcium transients, suggesting that E9.5 cranial neural crest cells are electrically coupled (Fig. 7A–E). At later stages, palate mesenchyme cells are dissociated before they are cultured, but they may form connections again in cultured conditions. To determine whether cells at E13.5 may also be electrically coupled, we performed network analysis where the Pearson’s Correlation Coefficients was first determined for all possible cell pairs throughout the time course, and cell pairs with a correlation >0.5 were considered ‘functionally connected’ or synchronized (Fig. 7G–L). The synchronized activity we observed suggests an electrical coupling between pairs of neighboring cells (Fig. 7F–L). Analysis of individual time traces for regions showing the highest degree of coordinated activity indicates some synchronized activity even in far-apart cells (Fig. 7L). These data further suggest that cranial neural crest cells and palate mesenchyme cells are electrically coupled.
A A representative image shows a cultured E9.5 explant with three representative cells manually selected threshold of 0.5. B Normalized calcium time trace of highly connected cells exhibiting a correlation coefficient > 60% of average connections across all cells. C Percentage of the area that shows highly correlated calcium activity within each field of view for 24-h (n = 4 independent E9.5 explants) vs 48-h incubated explants (n = 3 E9.5 independent explants, unpaired two-tailed Student t test, P = 0.3630). Error bars represent SEM. D False color map represents elevated calcium response time of selected cells in a cultured E9.5 explant with more activity. E Corresponding calcium time trace of cells highlighted in (D) with purple curves corresponding to cells initiating the observed calcium activity and pink curves corresponding to slower activating cells. F Scatter plot comparing Euclidean pixel distance between cell nuclei and Pearson correlation coefficient showing a general decrease in correlation with increasing distance between cells (G, J) Cell maps of E13.5 palate mesenchyme cells manually selected ROIs from which calcium time traces were extracted. H, K Highly connected cells exhibiting a correlation coefficient >0.5 are indicated in yellow on Connection Maps. I, L Example calcium time traces exhibit instances of synchronized activity by highly connected cell pairs. M, N, P, Q Confluent cells were hand-selected as a region of interest from which the fluorescent calcium signal was extracted and analyzed on a pixel-by-pixel basis to obtain peak amplitude values among individual pixel time courses. O, R Individual calcium signal time courses are extracted for individual cells of regions showing the highest intensity peak amplitude. Individual cell activity was greater in untreated samples compared to Carbenoxolone (CBX) treated as indicated by observed spike-like behavior shown in the signal. S The number of high-amplitude events was quantified for each sample. Untreated explants showed greater high-amplitude events compared to CBX treated (N = 4 CBX treated explants and 10 control explants unpaired two-tailed Student t test, P = 0.0180) Error bars represent SEM. T Area active was determined as a proportion of pixel area that was both correlated and showed fluorescence above background signal. Explants treated with gap junction inhibitor CBX showed a decrease in area active compared to untreated (N = 4 CBX treated explants and 10 control explants unpaired two-tailed Student t test, P = 0.0258). Error bars represent SEM.
Gap junction inhibition reduces calcium activity
To determine if gap junctions contribute to the spreading of calcium between cranial neural crest cells, we measured how correlated calcium transients were, in the presence and absence of pharmacological gap junction inhibition. Carbenoxolone (CBX) is a widely used broad-spectrum gap junction antagonist. We used CBX to inhibit gap junctions in E9.5 explant cultures. CBX inhibition did not affect cell or explant viability, as shown by repeated experiments where CBX was washed off, explants cultured overnight, and imaged subsequent days. Explants were analyzed for high-amplitude calcium transients, and the Pearson’s Correlation was calculated to assess for the effect of gap junction inhibition. We specifically tested whether instances of high-amplitude calcium transients decreased with gap junction inhibition via CBX (Fig. 7M–T). Indeed, cells showed significantly fewer high-amplitude calcium transients when treated with a gap junction inhibitor compared to untreated samples (Fig. 7R, S). Thus suggests that gap junctions contribute to the generation of high-amplitude calcium transients (as indicated by high-amplitude changes in fluorescence). Next, we asked whether the synchronization of calcium transients decreased when gap junctions were inhibited. Gap junction inhibition decreased the area of the explant exhibiting correlated activity compared to that measured under control conditions (Fig. 7O, R, T). When comparing clusters of cells and the surrounding regions showing high-amplitude transients, untreated explant cells showed more spiking or single oscillatory behavior compared to CBX treated (Fig. 7M–O, S). These results indicate a decrease in synchronized calcium activity area and reduction in high-amplitude calcium transients in confluent explant tissue treated with gap junction inhibitors (Fig. 7S, T). These data support the possible role of gap junctions in explant tissue facilitating the accumulation of calcium and subsequent release of BMP.
Ion channels are expressed throughout the palatal mesenchyme
To determine which ion channels are expressed in the palate mesenchyme and thus could regulate intracellular calcium at a time when BMP4 signaling is active, we identified ion channels that are expressed in an E13.5 anterior palate single-cell RNA sequencing (scRNAseq) dataset produced in our lab51. Mesenchymal and epithelial cell populations were identified by marker gene expression51. We identified the expression of ion channel-encoding genes previously associated with craniofacial defects (Cacna1a and Cacna1c: CaV1.2, Kcnj2: Kir2.1, and Gja1: Cx43, Fig. 8). Excitingly, we detected expression of ion channels that could regulate depolarization events in the anterior palate mesenchyme. We were particularly interested to note that voltage-gated sodium channels (Scn3a: NaV1.3, Scn8a: NaV1.6), voltage-gated calcium channels (Cacna1d: CaV1.3, Cacna1g: CaV3.1), potassium channels (Kcnb1: Kv2.1, Kcnb2: Kv2.2, Kcnc3: Kv3.3, Kcnn2: SK2), endoplasmic reticulum regulatory proteins (Atp2a2: SERCA2, Stim1, Stim2), and gap junction proteins (Gja1 Cxn43 and Gjc1: Cxn45) (Fig. 8) are expressed in the mouse E13.5 anterior palate mesenchyme and epithelial cells. Many other calcium, potassium, and sodium channels are expressed in the mesenchymal and epithelial cells that could contribute to regulating endogenous electrical activity in the anterior palate at E13.5 (Supplemental Figs. 5–7). It is important to note that ion channels can affect membrane potential even at low expression levels. scRNAseq at one embryonic time point likely does not detect every channel that contributes to craniofacial development. Kcnj2 is not highly expressed in the same palatal mesenchyme cells as BMP4 (Fig. 8R–T). However, virtually every cell in the anterior palate expresses at least one gene encoding gap junction proteins (Fig. 8U–W). With our observations that calcium transients are often synchronous between multiple cells, ubiquitous gap junction expression in the anterior palate suggests that these cells are electrically coupled and can propagate bioelectrical signals across long distances.
A UMAP detailing cluster identities adapted from Ozekin et al.51. FeaturePlots represent data from a single-cell RNA sequencing of the E13.5 mouse anterior palate showing expression of ion channels and connexins in green with non-expressing cells in gray: B Cacna1c (Cav1.2, L-type calcium channel). C Cacna1d (Cav1.3, L-type calcium channel). D Cacna1a (Cav1.2, L-type calcium channel). E Cacna1g (T-type calcium channel). F Kcnj2 (Kir2.1, inwardly rectifying potassium channel). G Kcnb1 (Kv2.1, voltage-gated potassium channel subfamily B). H Kcnb2 (Kv2.2), I Kcnc3 (Kv3.3, voltage-gated potassium channel subfamily C), J Kcnn2 (KCa2.2, Potassium Calcium-activated channel subfamily N), K ATP2a2 (SERCA2/Atpase Sarcoplasmic/Endoplasmic Reticulum Ca2+ Transporting 2), L Stim1, M Stim2, N Scn3a (Nav1.3, voltage-gated sodium channel, O Scn8a (Nav1.6 Voltage-gated sodium channel), P Gja1 (Gap junction protein alpha, Connexin 43), Q Gjc1(Gap Junction protein gamma 1, Connexin 45), R–T FeaturePlots of Bmp4 (Bone morphogenetic protein 4) (green), Kcnj2 (orange), and overlapped FeaturePlot of Bmp4 and Kcnj2. Cells with high coexpression of both features will appear on a gradient to yellow. U–W FeaturePlots of Gjc1 (green), Gja1 (orange), and overlapped FeaturePlot of Gjc1 and Gja1. Cells with high coexpression of both features will appear on a gradient to yellow. RNA sequencing data is available at Raw and processed data has been made available via a NCBI GEO Submission (accession code GSE222205). Code is accessible via GitHub https://github.com/yunusozekin/WT_E13.5_AntPalate_scRNAseq_Ozekin.git.
Discussion
Here, we provide evidence that depolarization induces BMP release from mouse palatal mesenchyme cells. We show depolarization-induced BMP release is dependent on cytoplasmic calcium. Furthermore, we show that palatal mesenchyme cells undergo transient changes in intracellular calcium, regulated by Kcnj2, a potassium channel required for palate development in mice and humans. This work suggests a mechanism linking observed craniofacial phenotypes of channelopathy patients with disruptions in traditional morphogen signaling. The necessity of ion channels for proper morphogenesis of human structures has been repeatedly documented (KCNJ2: Anderson-Tawil syndrome, CACNA1C: Timothy Syndrome, GIRK2: Keppen-Ludinski syndrome, NALCN: IHPRF1, and CHRNA7: 15q13.3 microdeletion syndrome)8,9,12,13,15,29,52,53,54. Similarly, pharmacological and genetic disruption of ion channels during embryonic development causes morphological abnormalities in animal models55. We propose that changes in membrane potential control secretion of vesicle-contained BMP ligands from palatal mesenchyme, in a calcium-dependent manner for craniofacial development.
Craniofacial phenotypes are not specific to the disruption of one type of ion channel, suggesting that electrical activity mediated by several ion channels contributes to developmental signaling. Genetic and clinical evidence indicates that ion channels are important for BMP signaling26,27,28. Mutations that disrupt Kcnj2 cause craniofacial abnormalities such as dental defects, cleft lip and palate, hypertelorism, low-set ears, and micrognathia (small jaw) in human Andersen-Tawil syndrome (ATS) patients and mouse knockouts9,13,26,28,32,56. BMP signaling is required for the development of the structures affected in ATS patients. Kcnj2 knockout (Kcnj2KO/KO) mice have similar craniofacial defects as ATS patients and as mice with disrupted BMP signaling26,28,48,57,58,59. Deletion of Kcnj2 specifically in the cranial CNC causes similar craniofacial phenotypes as deletion of the BMP receptor - BMPR1a in the CNC57. E13.5 Kcnj2KO/KO palatal shelves have decreased phosphorylation of Smad 1/5/8, a downstream target of BMP signaling and reduced BMP target gene expression26. In the developing Drosophila wing disc, depolarization induces BMP/Dpp release27. Our work suggests one possible conserved mechanism by which ion channels coordinate developmental signaling is by controlling release of BMP4.
We developed a novel tool to visualize BMP4 release from cells using the pH-sensitive GFP variant, super ecliptic pHluorin (SEP). The BMP4-SEP release reporter is useful for the BMP research field enabling visualization of BMP release events. In some cells, we observe a slight increase in fluorescence in pcDNA-SEP expressing cells upon depolarization potentially due to small changes in cytoplasmic pH. In comparison, both the TFR-SEP and the BMP4-SEP underwent more profound increases in fluorescence in the presence of KCl. TFR-SEP is localized to vesicles and is commonly used as a marker of vesicular release, demonstrating that iMEPM cells, derived from the palate shelves of E13.5 mice60, contain the proper machinery required for depolarization-induced vesicular fusion. Furthermore, we conclude that BMP4 is released from vesicles upon depolarization. The punctate appearance of TFR-SEP and BMP4-SEP fluorescence post-depolarization is consistent with vesicular fusion to the membrane. Future research is needed to determine the types of vesicles that contain BMP ligands and the specific mechanisms of release.
In excitable cells, such as neurons, if depolarization reaches a threshold at which voltage-gated Ca2+ channels open, increased Ca2+ levels change conformation of Soluble N-ethylmaleimide-Sensitive Factor Attachment Proteins REceptor (SNARE) proteins to induce vesicular fusion with the cellular membrane61. If BMP-containing vesicle fusion is regulated by the same SNARE-dependent mechanism, sequestering intracellular Ca2+ would prevent an increase in BMP4-SEP fluorescence upon depolarization. A calcium chelator, BAPTA-AM, prevented any increase in BMP4-SEP fluorescence upon depolarization in most cultured palate mesenchyme cells. There were small increases in BMP4-SEP fluorescence upon depolarization in two BAPTA-AM-treated cells which were significantly reduced in amplitude compared to cells that were not treated with BAPTA-AM. This suggests that BAPTA-AM treatment reduced the number of vesicle fusion events upon depolarization. Interestingly, in Drosophila, mutations in components of the SNARE protein complex have developmental defects62,63,64. Perhaps there is a conserved role for calcium and SNAREs in morphogenesis.
Cells use chemo-transduction to instruct transcription in neighboring cells at a designated point in morphogenesis. In early murine development, Wingless-related integration sites (Wnts), Bone Morphogenetic Proteins (BMPs), Sonic hedgehog (Shhs), Fibroblast Growth Factors (FGFs) and other pathways initiate signaling cascades that ultimately affect transcription with cells to influence cell fate and ultimately morphogenesis65. The temporal pattern of signaling affects the downstream transcriptional output in cell culture and in zebrafish osteoblast regeneration45,46. How do cells in developing tissues regulate these pathways to mediate the precise communication required for developmental decisions? Neurons, which also need precise communication, use depolarization as a critical step for vesicular release to regulate the release of molecular signals. We discovered cranial neural crest cells and palate mesenchyme cells, which are precursors to the palate, undergo rapid changes in intracellular calcium that are often cyclic and periodic, reminiscent of calcium spikes in neurons. Calcium transients are present in the neural crest precursor cells at E9.5, before BMP4 is expressed for palatogenesis. This raises the possibility that calcium transients may coordinate other signals in addition to BMP. Ion channel disruption affects other developmental pathways like Notch66,67,68, Shh69, and Wnt70,71,72,73. Our work opens the question of whether electrical activity affects the extracellular presentation of these essential developmental signaling ligands as it does for BMP4.
We observed groups of cells undergo coordinated calcium transients suggesting that the cells are electrically coupled via gap junctions (Figs. 6 and 7). Electrical coupling would allow groups of cells to temporally coordinate periodic waves of BMP release across distances. Indeed, we found that gap junction genes are ubiquitously expressed within the E13.5 palatal mesenchyme (Fig. 8U–W). The expression of one of these gap junctions, Gja1 (Cnx43), overlaps with the expression of Kcnj2 (Fig. 8S, U, W). Genetic disruption of Gja1 causes craniofacial phenotypes like that of Kcnj2 perturbation74. Gja1 modulates TGF-β1 and BMP2/4-mediated ERK signaling75. ERK, a downstream target of BMP signaling, relies on periodic waves of activation to control osteoblast differentiation and regeneration of scales in zebrafish46. Our data show that inhibition of gap junctions in E9.5 palatal explants significantly reduces calcium activity in both active area and number of high-amplitude calcium events (Fig. 7S, T). These results provide the basis for a mechanism explaining this observed temporal coordination of ERK signaling. In Xenopus laevis and Danio rerio, ion channels and gap junctions contribute to patterning and development suggesting that mechanisms controlling BMP ligand release could be further conserved25,31,76,77,78. Work from the Taneyhill laboratory supports the hypothesis that gap junctions between cranial neural crest cells are important for craniofacial development79,80. Cnx43/Gja1 is expressed in cranial neural crest cells in chick embryos79. Cnx43/Gja1 is required in cranial neural crest cells for their migration80. Our data provides adds another potential explanation for why mutations in gap junction genes cause craniofacial defects in humans81,82,83,84,85,86.
The difference in BMP4 and Kcnj2 expression could be interpreted in multiple ways. We favor the possibility that gap junctions connect cells within the palate so that ions spread throughout these cells to coordinate synchronous release of BMP. Another possibility is that RNA transcripts do not reflect Kcnj2 protein present in the cells. Ion channels are notoriously lowly expressed and can affect membrane potential even at low expression levels. Thus, scRNAseq can be used to deduce expression of a channel-encoding gene, but not the absence of a channel. A third possibility is that inwardly rectifying K+ channels encoded by different genes work together to set membrane potential and coordinate release of BMP ligands. Inwardly rectifying K+ channels are composed of four subunits and can be heteromeric. For example, we detect expression of Kcnj2, Kcnj3, Kcnj4, Kcnj6, Kcnj8, Kcnj9, Kcnj13, Kcnj14, Kcnj15, and Kcnj16 in our scRNAseq E13.5 anterior palate dataset (Supplemental Fig. 6). These inwardly rectifying channels could regulate membrane potential with overlapping function. Many other potassium channels are also expressed and likely aid in the recovery from depolarization events. Future research will address how multiple ion channels work together to coordinate endogenous changes in membrane potential, calcium levels, and ultimately release of BMP ligands.
How depolarization events are initiated and regulated within the palate mesenchyme is an outstanding question. Our scRNAseq dataset shows that a multitude of ion channels are expressed in the mesenchyme and epithelia of the mouse E13.5 palate. Future studies will determine if voltage-gated sodium and calcium channels initiate and regulate endogenous depolarization events. For example, we could determine if voltage-gated channels are required for calcium oscillations using pharmacological or genetic tools.
We hypothesized that ion channels that are important for craniofacial development regulate palatal intracellular calcium transients. Loss of one or both copies of Kcnj2 caused an overall decrease in GCaMP fluorescence, suggesting that basal calcium levels are reduced within these cells. Also, loss of Kcnj2 resulted in a significant decrease in calcium transient amplitude in palatal and CNC cells. Our data supports a model in which ion channels control membrane potential to mediate the secretion of BMP4. When ion channels are impaired, cells that rely upon those channels cannot regulate membrane potential, which disrupts intracellular calcium transients, propagation of these calcium signals, and consequently BMP secretion. Lack of regulated BMP secretion disrupts morphogenesis of the craniofacial complex (Fig. 9). This model explains a potential mechanism by which individuals with channelopathies have abnormal morphological development.
In wild-type cells, ion channels regulate depolarization events to control pulsatile intracellular calcium activity. Calcium drives fusion of BMP-containing vesicles allowing for the release of BMP ligands. In channelopathy cells, depolarization events are disrupted, resulting in decreased calcium activity and altered fusion of BMP vesicles to yield dysregulated release of BMP ligands.
Depolarization likely alters development through multiple mechanisms. In neurons, depolarization regulates release of neurotransmitters, but also induces different gene expression programs by calcium activating MAPK/ERK and CREB-mediated transcription87. In fact, different temporal patterns of depolarization induce different transcriptional profiles87. Calcium also influences chromatin structure to affect transcriptional profiles88,89. Depolarization likely impacts cells that send developmental signals by multiple mechanisms as well. Brief pulses of calcium could activate calcium signaling cascades to influence chromatin structure, transcription factor activity, and ultimately the expression of genes that are important for proliferation, differentiation, and migration.
This research has clinical implications for fetal exposures to medications and recreational drugs that inhibit or activate ion channels. Fetal exposure to certain drugs that affect ion channel function increases the incidence of birth defects55. For example, fetal exposure to an epilepsy medication and migraine medication called Topiramate increases the incidence of cleft palate90. Topiramate blocks voltage-dependent Na+ channels and AMPA glutamate receptors91,92,93. Similarly, pentobarbital use during the first trimester increases incidence of cleft lip/palate in humans and in rodents and blocks GABAA receptors, which are ligand-gated Cl- ion channels19,94. In addition, Trimethadione, which blocks voltage-dependent T-type Ca++ channels, significantly increases the incidence of cleft palate, ear defects, and limb defects in the children of women who took the medication during the first trimester of pregnancy18,95,96. We identify many previously uncharacterized ion channels expressed within the palatal mesenchyme that are targets of Pharmacologics (Fig. 8). Special care should be taken to understand how perturbation of these channels could affect palatal and craniofacial development. It may be that exposure to these drugs in utero disrupts cellular membrane potential to effect release of key morphogens, such as BMPs, thereby resulting in craniofacial defects.
Limitations of study
We show that BMP4 release can be induced by depolarization in cultured IMEPM cells and that primary cranial neural crest and palate mesenchyme cells undergo transient changes in intracellular calcium. Our assessment of depolarization-induced BMP secretion was performed in vitro and ex vivo, and this mechanism has not been confirmed in vivo. While Kcnj2KO/KO mouse data are consistent with our conclusions, direct assessment of depolarization and BMP secretion in vivo are not currently feasible. Furthermore, our studies do not allow us to draw conclusions about other mechanisms by which loss of Kcnj2 function could impact BMP signaling or other developmental signaling pathways. It is possible that membrane potential could affect BMP stability, which we have not addressed. Furthermore, changes in cytoplasmic calcium levels likely affect gene expression cell-autonomously through calcium-sensitive transcription factors or chromatin remodelers. Therefore, changing membrane potential could increase the expression of BMP inhibitors as a second possible mechanism by which Kcnj2 and other ion channels could impact BMP signaling. Our results do not exclude the possibility that multiple mechanisms work together to decrease BMP signaling and disrupt craniofacial development in Kcnj2KO/KO mice.
Ion channels are often lowly expressed and can impact membrane potential with low level expression. Thus, scRNAseq does not have the sensitivity to definitively state the absence of a channel, but rather can comment on those that can be detected. Furthermore, our scRNAseq is a snapshot of ion channel expression in the anterior palate at E13.5. Other ion channels are likely expressed and mediate membrane potential at different stages of palatal development.
We concluded that gap junctions connect cranial neural crest cells and dissociated palate mesenchyme cells because of the expression of gap junction encoding genes and quantitative analysis of synchronized changes in GCaMP fluorescence. This conclusion could be supported with paired electrophysiological recording in which current is injected in one cell and recorded in a neighboring cell97,98,99. Another method to determine whether cells are coupled by gap junctions is to trace coupling ion sensitive dyes or other dyes using wide-field imaging98. These rigorous assays will further characterize electrical coupling.
Methods
Mouse husbandry and approvals
All protocols and procedures were approved by the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee (IACUC) committee (protocol #139). The University of Colorado, Office of Laboratory Animal Research (OLAR) oversees an Assessment and Accreditation of Laboratory Animal Care accredited animal facility that meets standards as outlined in United States National Institute of Health Guide for the Care and Use of Laboratory Animals. Mus Musculus (mice) were fed a standard chow diet. Kcnj2KO/+ mice were originally purchased from Jackson Laboratories (FVB.129-Kcnj2tm1Swz/J strain #005057)32. These were bred within our facility to maintain a colony. Wnt-1 Cre mice are B6.Cg-E2f1Tg(Wnt1-cre)2Sor/J (#:022501). GCamp6 mice are from Jackson labs (stock number Ai96 GCaMP6s, #024106).
BMP4-SEP cloning strategy
To generate a reporter of BMP4 release, we tagged BMP4 with super ecliptic pHluorin (SEP), a pH-sensitive GFP. To retain proper protein structure and function, SEP was inserted into the linker domain of BMP4 (RRKKNKN– SEP–CRRHSLYVDFSD), as previously described in D. melanogaster27. The BMP4-SEP construct was generated for our use by GenScript. The BMP4 feature of the plasmid pDONR233_BMP4_WT_V5, purchased from Addgene (Catalog #: 82937), was cloned into the pcDNA3.1(+) vector with a CMV promotor. A gBlock of SEP purchased from and generated by Twist Biosciences in San Francisco, California was sub-cloned into the linker domain described above. We confirmed the construct’s sequence using DNA sequencing through Quintara Biosciences.
pcDNA3.1-SEP cloning strategy
A control plasmid for depolarization imaging was generated using SEP driven by a CMV promotor. The plasmid pcDNA3.1(+) (donated to us by a colleague) was digested with restriction enzymes HindIII and EcoRI to generate a linear plasmid. The plasmid TfR-mCherry-SEP (donated to us by Dr. Kennedy’s lab at the University of Colorado-AMC) was amplified via PCR using primers HindIII-SEP (CAGaagcttATGAGTAAAGGAGAAGAAC, purchased from IDT) and EcoRI-SEP (TCGgaattcTTATTTGTATAGTTCATCCA, purchased from IDT) to generate a SEP sequence with a HindIII overhang on the 5’ end and a EcoRI overhang on the 3’ end. The HindIII-SEP-EcoRI product and linearized pcDNA3.1(+) plasmid were then ligated together using the standard protocol from NEB T4 DNA Ligase (1:5 vector to insert ratio; purchased from NEB, catalog #: M0202S). The construct sequence was verified by DNA sequencing from Quintara Biosciences.
iMEPM cultures
iMEPM cells (immortalized mouse embryonic palatal mesenchyme) were cultured under the same conditions for each experiment. iMEPM cells were generated and donated by Dr. Fantauzzo’s lab at the University of Colorado, AMC and maintained by our lab. The media (referred to as iMEPM media from this point) used to grow the cells contained “DMEM with high glucose, no pyruvate or l-glutamine” (Gibco brand, purchased from ThermoFisher Catalog #: 11960044), 10% FBS (Gibco brand purchased from ThermoFisher catalog #: 16000044), 1% 200 mM l-Glutamine and 0.05% 50 mg/µl Pen Strep. The cells were passaged several times post-thawing and before splitting and plating for experiments. For depolarization imaging experiments, cells were cultured on WillCo Wells WillCo-dish® 35-mm glass bottom dishes (catalog #: HBST-3522) in 2 mL of standard iMEPM media (@37 °C; 5% CO2). For the ELISA experiments, iMEPM cells were cultured in 35 mm six-well plates in standard iMEPM media (@37 °C; 5% CO2).
MEF isolation and maintenance
Heterozygous Kcnj2ko/+ mice were mated, and dams were considered E0.5 on the day a vaginal plug was observed. At E14.5, embryos were harvested. Heads were removed for phenotypic analysis. Internal organs were isolated for genotyping. Each embryo was dissociated with 3 mL of. 0.25% trypsin in EDTA and at 37 °C for 5 minutes. Dissociated cells from each embryo were plated separately. We added 5 mL of a mixture of 10% DMEM, 1% FBS, 1% GlutaMAX (Gibco Life Technologies) and centrifuged for 5 min at 2000×g. The supernatant was replaced and resuspended in fresh 10% DMEM, 1% FBS, 1% GlutaMAX Pen Strep. Dissociated cells from each embryo were plated separately in six-well plates.
Quantitative RT-PCR
RNA was extracted from 75% confluent mouse embryonic fibroblasts in 10-cm dishes using a Promega RNA extraction Kit (catalog #73100). On the same day, RNA was reverse transcribed to cDNA using the Qiagen RT2 First Strand Kit (catalog # 330401) on a Roche lightcycler 480. We quantified cDNAs with SYBR green mastermix (Cat. #330502) and primers from a TGF-beta/BMP PCR array (catalog #330231) following the published protocol (95 °C for 10 min for heat activation of the enzyme, followed by PCR cycling for the acquisition of data using 45 cycles of 95 °C for 15 s followed by 60 °C for 1 min with the acquisition. Finally, the melt curve included 60 °C for 15 s followed by 95 °C with continuous acquisition of fluorescence signal. The genomic DNA control shows a peak past 35 cycles for all samples meaning that there is negligible genomic DNA contamination. We determined fold expression difference using the ddCT method. The mean of two technical and two biological replicates of wild-type and Kcnj2KO/KO replicates were normalized with the geometric mean of five housekeeping genes (Actb, B2m, Gapdh, Gusb, Hsp90ab1) to determine ΔCT = CTAVG GOI- CTAVG HKG. The standard deviation for ΔΔCT = CTAVG KO- CTAVG WT was carried over from the standard deviations calculated for the ΔCT KO samples100,101. Comparative analysis between WT and Kcnj2KO/KO expression (and standard deviation) uses the 2−ΔΔCT method to determine BMP fold difference in expression. To determine significance, a two-tailed, unpaired Student’s t test was performed using the ΔCT WT and Kcnj2KO/KO replicates for genes encoding each Bmp ligand. CT values for BMP3, BMP5, and BMP7 were above 35 in WT MEFs, indicating very low expression and thus were not reported in the main figure (see source file). Some CT values of BMP2, were above 35, indicating low expression (see source file).
Western blots
Protein isolated from WT cells treated with conditioned media was separated by gel electrophoresis and transferred to Turbo nitrocellulose transfer packs (BioRad). The blot was blocked for 1 h in 5% milk/TBST shaking at room temperature. Following blocking, the blot was incubated in 1:1000 Rabbit Phospho-Smad 1/5/8 S463/465, (Cell Signaling #13620) in 5% milk/TBST shaking in 4 °C overnight. The loading control was Rabbit GAPDH Cell Signaling Technologies (Cat#:2118). The blot was incubated in 1:2000 secondary anti-rabbit Goat Anti-(Rabbit IgG (H + L) Alexa Fluor Plus 555 Invitrogen Cat#:A32732) for 1 h at room temperature in the dark. The blot was washed four times in TBST for 15 min at room temperature before imaging with a BioRad Chemidoc MP Imaging System. Protein from three wild-type MEF plates treated with WT-conditioned media was loaded next to protein from three WT MEF plates treated with Kcnj2KO/+ conditioned media from three independent embryos, followed by three WT MEF plates treated with conditioned media from three independent Kcnj2KO/KO embryos in the representative western blot shown in Fig. 1. This was repeated in a separate western blot with the exception that there were only two independent Kcnj2KO/+ MEF samples. Thus N = 6 WT MEF conditioned media samples, N = 5 independent Kcnj2KO/+ MEF conditioned media, and N = 6 independent Kcnj2KO/KO MEF conditioned media. These two full western blots are provided in the accompanying source data file.
Transfections
To examine the release of BMP4 from cells, we co-transfected iMEPM cells with either the BMP4-SEP construct and mCherry or pcDNA3.1-SEP and mCherry. Transfections followed the standard “Lipofectamine® LTX & PLUS™ Reagent” kit protocol (purchased from ThermoFisher, catalog #: 15338030). iMEPM cells were cultured in six-well plates for ELISA experiments and on WillCo Wells WillCo-dish® 35-mm glass bottom dishes (catalog #: HBST-3522) for the depolarization imaging experiments. Cells were cultured until 80–90% confluent in standard iMEPM media, and media was then replaced with transfection solution. 1 mg of each type of DNA (BMP4-SEP and mCherry or PCDNA3.1-SEP and mCherry) was diluted in “Opti-MEM™ Reduced Serum Medium with no phenol red” (catalog #: 11058021) along with PLUS reagent and Lipofectamine-LTX from the transfection kit above. The solution was added to each plate dropwise and allowed to incubate at 37 °C; 5% CO2 for 4 h, the solution was then replaced with iMEPM culture media and allowed to incubate overnight before imaging/ extraction (24 h post transfection).
Imaging and data analysis
Depolarization imaging was performed using a Zeiss LSM 880 confocal microscope with Airyscan. The images were taken as a 200 cycle time series at a speed of 250 ms with an average Airyscan scan time of 248.66 ms. To capture the SEP fluorescence, cells were excited at 488 nm. To reduce file size while optimizing resolution, files were Airyscan processed after recording. Cells were imaged in iMEPM media. To capture the depolarization of the iMEPM cells, a solution of KCl (50 mM, based on the neuroscience field standard concentration for cell depolarization102,103,104,105 was added to the dish via pipette at frame 50 while imaging. SEP fluorescence was measured before and after the depolarization event using Fiji, and amplitude was calculated for each event observed. Statistical analysis was done using the Student’s t test function in GraphPad Prism8 to determine the significance of the difference in fluorescence.
Isosmotic depolarization in BAPTA
Isosmotic depolarization and BAPTA-AM (ThermoFisher Scientific, #B1205) imaging was performed using a Zeiss LSM 900 confocal microscope with Airyscan II using the 40× water objective. Capture parameters were the same as the depolarization methods detailed above. To cause depolarization under isosmotic conditions, cells were imaged in a low potassium, high sodium Tyrode’s media (135 mM NaCl, 5 mM KCl, 2 mM CaCl2·2H2O, 1 mM MgCl2·6H2O, 25 mM HEPES, 10 mM Glucose, and 0.10% BSA, pH 7.4 with NaOH). At 150 frames, a high potassium, low sodium Tyrode’s media (5 mM NaCl, 135 mM KCl, 2 mM CaCl2·2H2O, 1 mM MgCl2·6H2O, 25 mM HEPES, 10 mM Glucose, and 0.10% BSA, pH 7.4 with NaOH) was added directly to the plate to bring a the KCl to a final concentration of 50 mM. Cells were exposed to either 100 mM BAPTA-AM diluted in DMSO and Tyrode’s media or DMSO in Tyrode’s media alone for one hour at 37 °C prior to imaging.
ELISA and analysis
A BMP4-Mouse ELISA assay was performed to chemically confirm the presence of BMP4 in the ell media before and after depolarization. Cells were cultured and transfected with BMP4-SEP and mCherry as described above were depolarized with KCl (50 mM). Fractions of media were collected before and after depolarization was induced. Fractions were prepared and assayed according to the standard protocol from the Abnova “BMP4 (Mouse) ELISA Kit” (catalog #: KA5051). The ELISA plate was read on the Biotek synergy H1microplate reader at 561 nm. Statistical Analysis was performed using the Students’ t test function in GraphPad Prism8 to determine the significance of the difference in fluorescence intensity before and after depolarization.
Crystal violet assay and analysis
We followed the Cold Springs Harbor 2016 “Crystal violet assay for determining the viability of cultured cells” protocol (M. Feoktistova, P. Geserick, and M. Leverkus). Briefly, iMEPM cells were seeded onto 96well plates, leaving at least three wells with no cells (media only). Cells were cultured overnight. Each KCl concentration was added to treatment wells in triplicate, leaving three seeded cells untreated as a negative control. Three wells were also treated with H2O2 to induce cell death as a positive control. KCl treatment was applied for 15 min to simulate experimental conditions of depolarization assays. The wells were then washed and stained with 0.5% crystal violet solution. Prior to imaging, the wells were washed and air-dried overnight. Optical density was measured at 570 nm. Optical density values were graphed and used to calculate the percentage of light transmitted through the sample.
GCaMP methods
Rosa26-GCaMP6s (Jackson Laboratory, B6;129S6-Gt (ROSA)26Sortm96(CAG-GCaMP6s)Hze/J) female mice were mated overnight with WWnt1CRE males. Observation of a vaginal plug was considered day 0.5. On day 13.5, pregnant females were euthanized by isoflurane exposure and cervical dislocation. Embryos were dissected in ice-cold phosphate-buffered saline (PBS) and limbs were removed for genotyping.
Cell culture
Mouse embryonic palate (MEP) cells were collected from E13.5 embryos as per protocol adapted from (Bush and Soriano106). Briefly, paired palatal shelves were isolated from each embryo and placed in 100 µL 0.25% trypsin/EDTA (ThermoFisher Scientific, #25200056) for 15 min at room temperature with frequent agitation. Trypsinization was stopped by adding 10 volumes of media (DMEM + 10% FBS). Glass bottom culture dishes (Willco Wells, HBST-3522) were prepared by coating in 0.005% fibronectin in PBS for 45 min at room temperature immediately before dissection. MEP cells were plated and cultured overnight at 37 °C. The following day, cells were rinsed with PBS, and fresh media was added.
Embryonic day 9.5 neural tube explant dissection and culture
Neural tube explants were collected from E9.5 embryos by making incisions to separate the head and brain from the body, and one to separate the neural tube (NT) from the heart and ventral side of the body from the dorsal neural tube. NT explants are dissected at E9.5 while CNC cells are migrating ventrally to give rise to the palate and other craniofacial bone structures. After dissection, NT explants are plated on a fibronectin-coated imaging dish with 20 µL of warmed media (10% FBS and DMEM) to promote attachment. Explants are closely monitored for attachment to the plate and prevention of drying every 30 min after dissection. After explants are attached to the plate, we add 20 µL media dropwise and incubate for an additional hour. Explants are cultured overnight in 300 µL of media and imaged the following day. CNCs migrate out of the explant tissue and form an array of cells connected to the cultured explant. Explants and CNCs that remain adherent are then used for imaging experiments described in the body of the text: calcium live imaging, CBX gap junction inhibition, and depolarization studies.
Calcium imaging and quantification
E9.5 CNC cells were imaged 24 h post dissection on a ZEISS LSM 880 with airyscan confocal microscope using the 488 nm laser at ×20 magnification at 4 Hz. over 2.5 min. E13.5 palate primary cultured cells were imaged 24 h post dissection on a ZEISS LSM 900. GCaMP6s signal was observed using a 488 nm laser under ×40 magnification at 3.16 Hz. All cells that showed high variance were selected as regions of interest (ROIs). Mean fluorescence profiles were plotted for each ROI over 6 min. Mean fluorescence profiles were used to obtain event numbers, event amplitude, and interevent period. An event was defined as a fluorescent spike two or more standard deviations above the mean fluorescence. Interevent periods were determined by measuring the time between the peaks of two adjacent amplitudes. Event numbers were calculated by counting the number of events, as defined above, within a cell. For each sample, a minimum of five fields of view (FOVs) were imaged.
Quantification of GCaMP number of events, F0, amplitude, and interevent interval
Number of events were counted in each cell where activity was observed within a field of view (WT: N = 3, n = 198; Kcnj2KO/+: N = 4, n = 159; Kcnj2KO/KO: N = 4, n = 103). An event was defined as having a max fluorescence of two standard deviations or greater above mean fluorescence or the trace. Amplitudes were defined as the max peak fluorescence value of an event minus the minimum value before that peak (WT: N = 3, n = 633; Kcnj2KO/+: N = 4, n = 460; Kcnj2KO/KO: N = 4, n = 556). F0, or initial fluorescence, was determined by finding the minimum fluorescence value of the line trace (WT: N = 3, n = 199; Kcnj2KO/+: N = 4, n = 159; Kcnj2KO/KO: N = 4, n = 156). The interval between two events within the same cell was measured as the interevent period (WT: N = 3, n = 172; Kcnj2KO/+: N = 4, n = 237; Kcnj2KO/KO: N = 4, n = 208). For iMEPM experiments, the average change in fluorescence was not quantified due to differences in GCamp plasmid copy number.
Gap Junction inhibition calcium signal analysis (Fig. 7A–E, M–T)
Explants were imaged at 350 ms/frame and analyzed using MATLAB. Confluent cells were hand-selected and analyzed. Peak amplitude of fluorescence was identified for each pixel and plotted as a heatmap relative to the mean where yellow indicates high amplitude and blue corresponds with low. The number of high-amplitude events was quantified based on regions within the heatmap showing high peak amplitude. Average correlation was assessed on a pixel-by-pixel basis and superimposed with the peak amplitude map to obtain ratio of ‘active’ area with respect to the entire tissue area. Specific cell clusters were hand-selected and assessed based on areas of high peak amplitude whereby calcium signal was extracted using MATLAB and plotted to compare against CBX treated samples.
Network analysis
Calcium was analyzed using MATLAB scripts designed to identify individual cells with highly correlated activity (Fig. 7G–R). First, single cells were identified by hand. Individual calcium time traces were extracted and normalized. Pearson’s correlation coefficient was derived for all identified cell pairs. Pairs with a correlation >0.5 were considered “connected” and mapped as a Boolean matrix.
scRNA-sequencing analysis
UMAP and FeaturePlots were generated using Seurat on a publicly available dataset profiling the E13.5 anterior palatal mesenchyme55.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data generated in this study have been deposited in Figshare [https://doi.org/10.6084/m9.figshare.26045629]. The primary whole western blots from Fig. 1 are also provided in the Supplementary Information and Source Data file. Raw and processed RNA sequencing data is available on NCBI GEO under accession code GSE222205.
Code availability
Code is accessible via GitHub https://github.com/yunusozekin/WT_E13.5_AntPalate_scRNAseq_Ozekin.githttps://zenodo.org/records/13887375. The code used to analyze GCaMP fluorescence is available associated with this article.
References
Marks, M. L., Trippel, D. L. & Keating, M. T. Long QT syndrome associated with syndactyly identified in females. Am. J. Cardiol. 76, 744–745 (1995).
Splawski, I. et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc. Natl. Acad. Sci. USA 102, 8089–8096 (2005).
Splawski, I. et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 119, 19–31 (2004).
Barel, O. et al. Maternally inherited Birk Barel mental retardation dysmorphism syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am. J. Hum. Genet. 83, 193–199 (2008).
Basel-Vanagaite, L., Shaffer, L. & Chitayat, D. Keppen-Lubinsky syndrome: expanding the phenotype. Am. J. Med. Genet. A 149A, 1827–1829 (2009).
De Brasi, D., Brunetti-Pierri, N., Di Micco, P., Andria, G. & Sebastio, G. New syndrome with generalized lipodystrophy and a distinctive facial appearance: confirmation of Keppen-Lubinski syndrome? Am. J. Med. Genet. A 117A, 194–195 (2003).
Deeb, R., Veerapandiyan, A., Tawil, R. & Treidler, S. Variable penetrance of Andersen-Tawil syndrome in a family with a rare missense KCNJ2 mutation. Neurol. Genet. 4, e284 (2018).
Donaldson, M. R. et al. PIP2 binding residues of Kir2.1 are common targets of mutations causing Andersen syndrome. Neurology 60, 1811–1816 (2003).
Donaldson, M. R., Yoon, G., Fu, Y. H. & Ptacek, L. J. Andersen-Tawil syndrome: a model of clinical variability, pleiotropy, and genetic heterogeneity. Ann. Med. 36, 92–97 (2004).
Horvath, G. A. et al. Gain-of-function KCNJ6 mutation in a severe hyperkinetic movement disorder phenotype. Neuroscience 384, 152–164 (2018).
Masotti, A. et al. Keppen-Lubinsky syndrome is caused by mutations in the inwardly rectifying K+ channel encoded by KCNJ6. Am. J. Hum. Genet. 96, 295–300 (2015).
Plaster, N. M. et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell 105, 511–519 (2001).
Yoon, G. et al. Andersen-Tawil syndrome: prospective cohort analysis and expansion of the phenotype. Am. J. Med. Genet. A 140, 312–321 (2006).
Zhang, L. et al. Electrocardiographic features in Andersen-Tawil syndrome patients with KCNJ2 mutations: characteristic T-U-wave patterns predict the KCNJ2 genotype. Circulation 111, 2720–2726 (2005).
Al-Sayed, M. D. et al. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am. J. Hum. Genet. 93, 721–726 (2013).
Alwan, S. et al. Use of selective serotonin-reuptake inhibitors in pregnancy and the risk of birth defects. New Engl. J. Med. 356, 2684–2692 (2007).
Anderson, K. N. et al. Maternal use of specific antidepressant medications during early pregnancy and the risk of selected birth defects. JAMA Psychiatry 77, 1246–1255 (2020).
Feldman, G. L., Weaver, D. D. E. & Lovrien, W. The fetal trimethadione syndrome: report of an additional family and further delineation of this syndrome. Am. J. Dis. Child 131, 1389–1392 (1977).
Fritz, H., Muller, D. & Hess, R. Comparative study of the teratogenicity of phenobarbitone, diphenylhydantoin and carbamazepine in mice. Toxicology 6, 323–330 (1976).
Hu, H. et al. Prenatal nicotine exposure retards osteoclastogenesis and endochondral ossification in fetal long bones in rats. Toxicol. Lett. 295, 249–255 (2018).
Kennedy, A. E., Kandalam, S., Olivares-Navarrete, R. & Dickinson, A. J. G. E-cigarette aerosol exposure can cause craniofacial defects in Xenopus laevis embryos and mammalian neural crest cells. PLoS ONE 12, e0185729 (2017).
Mines, D. et al. Topiramate use in pregnancy and the birth prevalence of oral clefts. Pharmacoepidemiol. Drug Saf. 23, 1017–1025 (2014).
Wang, H. et al. Chloride channel ClC-3 promotion of osteogenic differentiation through Runx2. J. Cell Biochem. 111, 49–58 (2010).
Hutson, M. R. et al. Temperature-activated ion channels in neural crest cells confer maternal fever-associated birth defects. Sci. Signal 10, eaal4055 (2017).
Adams, D. S. et al. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J. Physiol. 594, 3245–3270 (2016).
Belus, M. T. et al. Kir2.1 is important for efficient BMP signaling in mammalian face development. Dev. Biol. 444, S297–S307 (2018).
Dahal, G. R., Pradhan, S. J. & Bates, E. A. Inwardly rectifying potassium channels influence Drosophila wing morphogenesis by regulating Dpp release. Development 144, 2771–2783 (2017).
Dahal, G. R. et al. An inwardly rectifying K+ channel is required for patterning. Development 139, 3653–3664 (2012).
George, L. F. et al. Ion channel contributions to wing development in Drosophila melanogaster. G3 9, 999–1008 (2019).
Levin, M. Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 184, 1971–1989 (2021).
Perathoner, S. et al. Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10, e1004080 (2014).
Zaritsky, J. J., Eckman, D. M., Wellman, G. C., Nelson, M. T. & Schwarz, T. L. Targeted disruption of Kir2.1 and Kir2.2 genes reveals the essential role of the inwardly rectifying K(+) current in K(+)-mediated vasodilation. Circ. Res. 87, 160–166 (2000).
Cordero, D. R. et al. Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. A 155A, 270–279 (2011).
Simoes-Costa, M. & Bronner, M. E. Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257 (2015).
Srinivasan, A. & Toh, Y. C. Human pluripotent stem cell-derived neural crest cells for tissue regeneration and disease modeling. Front. Mol. Neurosci. 12, 39 (2019).
Kanzler, B., Foreman, R. K., Labosky, P. A. & Mallo, M. BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development 127, 1095–1104 (2000).
Meinecke, L. et al. Modeling craniofacial development reveals spatiotemporal constraints on robust patterning of the mandibular arch. PLoS Comput. Biol. 14, e1006569 (2018).
Murillo-Rincon, A. P. & Kaucka, M. Insights into the complexity of craniofacial development from a cellular perspective. Front. Cell Dev. Biol. 8, 620735 (2020).
Letsou, A. et al. Drosophila Dpp signaling is mediated by the punt gene product: a dual ligand-binding type II receptor of the TGF beta receptor family. Cell 80, 899–908 (1995).
Ruberte, E., Marty, T., Nellen, D., Affolter, M. & Basler, K. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell 80, 889–897 (1995).
Sekelsky, J. J., Newfeld, S. J., Raftery, L. A., Chartoff, E. H. & Gelbart, W. M. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139, 1347–1358 (1995).
Nohe, A. et al. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 277, 5330–5338 (2002).
Wang, R. N. et al. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105 (2014).
Chen, G., Deng, C. & Li, Y. P. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 8, 272–288 (2012).
Sorre, B., Warmflash, A., Brivanlou, A. H. & Siggia, E. D. Encoding of temporal signals by the TGF-beta pathway and implications for embryonic patterning. Dev. Cell 30, 334–342 (2014).
De Simone, A. et al. Control of osteoblast regeneration by a train of Erk activity waves. Nature 590, 129–133 (2021).
Burke, K. J. Jr. & Bender, K. J. Modulation of ion channels in the axon: mechanisms and function. Front. Cell Neurosci. 13, 221 (2019).
Bonilla-Claudio, M. et al. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development 139, 709–719 (2012).
James, R. E. et al. Crimpy enables discrimination of presynaptic and postsynaptic pools of a BMP at the Drosophila neuromuscular junction. Dev. Cell 31, 586–598 (2014).
Oliveria, S. F., Dittmer, P. J., Youn, D. H., Dell’Acqua, M. L. & Sather, W. A. Localized calcineurin confers Ca2+-dependent inactivation on neuronal L-type Ca2+ channels. J. Neurosci. 32, 15328–15337 (2012).
Ozekin, Y. H., O’Rourke, R. & Bates, E. A. Single cell sequencing of the mouse anterior palate reveals mesenchymal heterogeneity. Dev. Dyn. 252, 713–727 (2023).
Chong, J. X. et al. De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am. J. Hum. Genet. 96, 462–473 (2015).
Hoppman-Chaney, N., Wain, K., Seger, P. R., Superneau, D. W. & Hodge, J. C. Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin. Genet. 83, 345–351 (2013).
Nilius, B. & Voets, T. The puzzle of TRPV4 channelopathies. EMBO Rep. 14, 152–163 (2013).
Ozekin, Y. H., Isner, T. & Bates, E. A. Ion channel contributions to morphological development: insights from the role of Kir2.1 in bone development. Front. Mol. Neurosci. 13, 99 (2020).
Villanueva, S. et al. Cleft palate, moderate lung developmental retardation and early postnatal lethality in mice deficient in the Kir7.1 inwardly rectifying K+ channel. PLoS ONE 10, e0139284 (2015).
Baek, J. A. et al. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev. Biol. 350, 520–531 (2011).
Bandyopadhyay, A. et al. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2, e216 (2006).
Dudas, M., Sridurongrit, S., Nagy, A., Okazaki, K. & Kaartinen, V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech. Dev. 121, 173–182 (2004).
Fantauzzo, K. A. & Soriano, P. Generation of an immortalized mouse embryonic palatal mesenchyme cell line. PLoS ONE 12, e0179078 (2017).
Chen, Y. A. & Scheller, R. H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2, 98–106 (2001).
Lagow, R. D. et al. Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol. 5, e72 (2007).
Schulze, K. L. & Bellen, H. J. Drosophila syntaxin is required for cell viability and may function in membrane formation and stabilization. Genetics 144, 1713–1724 (1996).
Stewart, B. A., Mohtashami, M., Zhou, L., Trimble, W. S. & Boulianne, G. L. SNARE-dependent signaling at the Drosophila wing margin. Dev. Biol. 234, 13–23 (2001).
Sagner, A. & Briscoe, J. Morphogen interpretation: concentration, time, competence, and signaling dynamics. Wiley Interdiscip Rev. Dev. Biol. 6, e271 (2017).
Periz, G. M. & Fortini, E. Ca(2+)-ATPase function is required for intracellular trafficking of the Notch receptor in Drosophila. EMBO J. 18, 5983–5993 (1999).
Roti, G. et al. Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer. Cancer Cell 23, 390–405 (2013).
Suisse, A. & Treisman, J. E. Reduced SERCA function preferentially affects Wnt signaling by retaining E-cadherin in the endoplasmic reticulum. Cell Rep. 26, 322–329.e323 (2019).
Emmons-Bell, M. & Hariharan, I. K. Membrane potential regulates Hedgehog signalling in the Drosophila wing imaginal disc. EMBO Rep. 22, e51861 (2021).
Muccioli, S. et al. From channels to canonical Wnt signaling: a pathological perspective. Int. J. Mol. Sci. 22, 4613 (2021).
Breuer, E. K. et al. Potassium channel activity controls breast cancer metastasis by affecting beta-catenin signaling. Cell Death Dis. 10, 180 (2019).
Costa, R. et al. Impaired mitochondrial ATP production downregulates Wnt signaling via ER stress induction. Cell Rep. 28, 1949–1960.e1946 (2019).
Thrasivoulou, C., Millar, M. & Ahmed, A. Activation of intracellular calcium by multiple Wnt ligands and translocation of beta-catenin into the nucleus: a convergent model of Wnt/Ca2+ and Wnt/beta-catenin pathways. J. Biol. Chem. 288, 35651–35659 (2013).
Lecanda, F. et al. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J. Cell Biol. 151, 931–944 (2000).
Yamada, A. et al. Connexin 43-mediated gap junction communication regulates ameloblast differentiation via ERK1/2 phosphorylation. Front. Physiol. 12, 748574 (2021).
Daane, J. M. et al. Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci. Rep. 8, 10391 (2018).
Lanni, J. S. et al. Integrated K+ channel and K+Cl- cotransporter functions are required for the coordination of size and proportion during development. Dev. Biol. 456, 164–178 (2019).
Osorio-Mendez, D. et al. Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. Proc. Natl. Acad. Sci. USA 119, e2200342119 (2022).
Jourdeuil, K. & Taneyhill, L. A. Spatiotemporal expression pattern of Connexin 43 during early chick embryogenesis. Gene Expr. Patterns 27, 67–75 (2018).
Jourdeuil, K. & Taneyhill, L. A. The gap junction protein connexin 43 controls multiple aspects of cranial neural crest cell development. J. Cell Sci. 133, jcs235440 (2020).
Essenfelder, G. M. et al. Connexin30 mutations responsible for hidrotic ectodermal dysplasia cause abnormal hemichannel activity. Hum. Mol. Genet. 13, 1703–1714 (2004).
Ishikawa, M. et al. Pannexin 3 and connexin 43 modulate skeletal development through their distinct functions and expression patterns. J. Cell Sci. 129, 1018–1030 (2016).
Ishikawa, M. & Yamada, Y. The role of pannexin 3 in bone biology. J. Dent. Res. 96, 372–379 (2017).
Liu, W. et al. Gap junction-mediated cell-to-cell communication in oral development and oral diseases: a concise review of research progress. Int. J. Oral. Sci. 12, 17 (2020).
Liu, W. J., Zhang, D. M., Zhou, X. D. & Xie, J. The role of connexins and pannexins in the cell communications of bone cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 51, 771–776 (2020).
Yin, J. et al. Role of connexin 43 in odontoblastic differentiation and structural maintenance in pulp damage repair. Int. J. Oral Sci. 13, 1 (2021).
Tyssowski, K. M. et al. Different neuronal activity patterns induce different gene expression programs. Neuron 98, 530–546 e511 (2018).
Sharma, A. et al. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc. Natl. Acad. Sci. USA 111, E4920–E4928 (2014).
Phengchat, R. et al. Calcium ions function as a booster of chromosome condensation. Sci. Rep. 6, 38281 (2016).
Margulis, A. V. et al. Use of topiramate in pregnancy and risk of oral clefts. Am. J. Obstet. Gynecol. 207, 405 e401–405 e407 (2012).
Chiossi, L., Negro, A., Capi, M., Lionetto, L. & Martelletti, P. Sodium channel antagonists for the treatment of migraine. Expert Opin. Pharmacother. 15, 1697–1706 (2014).
Sitges, M., Guarneros, A. & Nekrassov, V. Effects of carbamazepine, phenytoin, valproic acid, oxcarbazepine, lamotrigine, topiramate and vinpocetine on the presynaptic Ca2+ channel-mediated release of [3H]glutamate: comparison with the Na+ channel-mediated release. Neuropharmacology 53, 854–862 (2007).
White, H. S., Smith, M. D. & Wilcox, K. S. Mechanisms of action of antiepileptic drugs. Int. Rev. Neurobiol. 81, 85–110 (2007).
Yaffe, K. & Lowenstein, D. H. Prognostic factors of pentobarbital therapy for refractory generalized status epilepticus. Neurology 43, 895–900 (1993).
Barton, M. E., Eberle, E. L. & Shannon, H. E. The antihyperalgesic effects of the T-type calcium channel blockers ethosuximide, trimethadione, and mibefradil. Eur. J. Pharm. 521, 79–85 (2005).
Nelson, M. T., Todorovic, S. M. & Perez-Reyes, E. The role of T-type calcium channels in epilepsy and pain. Curr. Pharm. Des. 12, 2189–2197 (2006).
Ma, B. et al. Gap junction coupling confers isopotentiality on astrocyte syncytium. Glia 64, 214–226 (2016).
Stephan, J., Eitelmann, S. & Zhou, M. Approaches to study gap junctional coupling. Front. Cell Neurosci. 15, 640406 (2021).
Zhong, S. et al. Electrophysiological behavior of neonatal astrocytes in hippocampal stratum radiatum. Mol. Brain 9, 34 (2016).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408 (2001).
Rienecker, K. D. A., Poston, R. G. & Saha, R. N. Merits and limitations of studying neuronal depolarization-dependent processes using elevated external potassium. ASN Neuro 12, 1759091420974807 (2020).
Rienecker, K. D. A. et al. Mild membrane depolarization in neurons induces immediate early gene transcription and acutely subdues responses to a successive stimulus. J. Biol. Chem. 298, 102278 (2022).
Sertel, S. M. et al. Differences in synaptic vesicle pool behavior between male and female hippocampal cultured neurons. Sci. Rep. 11, 17374 (2021).
Wu, G. R., Jing, S., Momma, K. & Nakanishi, T. The effect of vitamin A on contraction of the ductus arteriosus in fetal rat. Pediatr. Res. 49, 747–754 (2001).
Lewis, A. E., Vasudevan, H. N., O’Neill, A. K., Soriano, P. & Bush, J. O. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 379, 229–234 (2013).
Bates, E. BioRender.com/m56f424 (2024).
Acknowledgements
We thank our colleague, Dr. Katherine Fantauzzo, for generously donating the iMEPM cells and Dr. Colleen Bartman for performing western blots. We thank Dr. Mark Dell’Acqua for insightful comments and suggestions. We are especially grateful for funding for this work from the National Institute of Dental and Craniofacial Research NIH-NIDCR-R01DE025311 to E.A.B. and National Science Foundation 1945916 to E.A.B. and NIH-T32GM141742-02S1 to Y.H.O. The Benninger laboratory gratefully acknowledges support from the NIH/NIDDK (R01DK106412, R01DK102950, R01DK140904 to R.K.P.B.) and the University of Colorado Diabetes Research Center (P30 DK116073).
Author information
Authors and Affiliations
Contributions
E.A.B. conceived and supervised experiments. T.J.I. generated the BMP4-SEP plasmid. Y.H.O., T.J.I., E.A.B., and M.L.F. performed BMP4-SEP experiments. T.J.I. conducted iMEPM GCaMP experiments. Y.H.O. performed CNC and palate primary culture endogenous GCaMP activity experiments. M.L.F. performed and analyzed depolarization-induced increases in GCaMP fluorescence in palate primary cultures. M.L.F. performed gap junction inhibition experiments in primary explant cultures and migrating CNCs. C.B. performed qRT-PCR of BMP ligands. C.H.L. analyzed waves of GCaMP fluorescence and gap junction experiments under the mentorship of RKPB. Y.H.O. analyzed our published single-cell-RNA sequencing of the E13.5 anterior palate. M.L.F. conducted assessments of cell viability after depolarization. M.L.F., T.J.I., Y.H.O., C.H.L., and E.A.B. created the figures and wrote the first drafts of the components of the manuscript. E.A.B., R.K.P.B., and Y.H.O. provided funding. All authors worked together in editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Follmer, M.L., Isner, T.J., Ozekin, Y.H. et al. Depolarization induces calcium-dependent BMP4 release from mouse embryonic palate mesenchymal cells. Nat Commun 15, 9806 (2024). https://doi.org/10.1038/s41467-024-53642-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53642-2