Abstract
Cerebral vasospasm (CVS) is an important contributor to delayed cerebral ischemia following aneurysmal subarachnoid hemorrhage (aSAH), leading to high morbidity and long-term disability. While several microRNAs (miRNAs) have been implicated in vasospasm, the underlying mechanisms for CVS remain poorly understood. Our study aims to identify miRNAs that may contribute to the development of CVS. Whole-blood samples were obtained during or outside of vasospasm from aSAH patients whose maximal vasospasm was moderate or severe. MiRNAs were isolated from serial whole-blood samples, and miRNA sequencing was performed. Differentially expressed miRNAs were identified and the expression levels in patients’ samples were verified using real-time qPCR. The biological functions of identified miRNA were evaluated in human brain endothelial cells (HBECs). MiRNA profiling revealed significant upregulation of miR-148b-3p in patients during CVS. We demonstrated that miR-148b-3p directly targeted and decreased the expression of ROCK1, affecting cell proliferation, migration, and invasion of HBECs through the ROCK-LIMK-Cofilin pathway. We propose that the upregulation of miRNA-148b-3p plays a role in the development of CVS by regulating actin cytoskeletal dynamics in HBECs, which is crucial for vascular function. Our study highlights miR-148b-3p as a potential diagnostic marker as well as therapeutic target for CVS following aSAH.
Similar content being viewed by others
Introduction
Delayed cerebral ischemia (DCI) frequently occurs as a serious complication after subarachnoid hemorrhage (SAH) caused by the rupture of cerebral aneurysms1,2. It is characterized by delayed neurological deterioration and/or cerebral infarction3. It is observed in approximately 30% of patients who survive the initial SAH4,5 and result in significantly increased morbidity6,7,8. Cerebral vasospasm (CVS), which is defined as arterial narrowing, has been identified as a major cause of DCI following SAH9,10. Although recent studies have implicated other potential contributors to DCI, including microthrombosis, cortical spreading depression, neuroinflammation and microcirculatory dysfunction3, cerebral vasospasm (CVS) severity remains significantly associated with DCI-related cerebral infarction following SAH11. Several studies have demonstrated involvement of factors such as endothelial dysfunction, loss of autoregulation, and systemic inflammatory reaction in the pathogenesis of vasospasm12,13,14. Vessels can be influenced by phenotypic switches that promote the migration, proliferation, and de-differentiation of human brain endothelial cells (HBECs), smooth muscle cells (SMCs), or both15,16. Previous studies have indicated that during CVS, SMCs undergo apoptosis and are involved in regulating endothelial cell loss and the erosion of the endothelial surface in cerebral arteries17,18. In addition, an aneurysmal SAH (aSAH) can upregulate pro-inflammatory cytokines such as IL-1, IL-6, and TNF-alpha, which lead to endothelial dysfunction, including reduced nitric oxide production, upregulated coagulation, and increased permeability in endothelial cells19,20. This damage decreases anticoagulant molecules and prostacyclin, promotes platelet adhesion, and enhances platelet–leukocyte–endothelial cell interactions21,22,23,24.
MicroRNAs (miRNAs) are single-stranded non-coding RNAs containing 22–25 nucleotides that regulate the translational inhibition or degradation of mRNAs25,26,27. Several miRNAs have been found to be differentially expressed in vascular tissues, suggesting that miRNAs may be involved in the dysregulation of vascular endothelial cells28,29. It has been reported that abnormal expression of miRNAs contributes to neurological diseases such as stroke, aneurysm, and vasospasm30,31,32,33. Several studies have also revealed that miRNAs regulate inflammatory responses in the central nervous system, and affect cerebral arteries following stroke and aSAH34,35,36,37. In addition, these miRNAs can be secreted into the blood38,39. Circulating miRNAs in the blood regulate inflammation, the interplay between inflammatory and vascular cells, and oxidative stress40,41. Several miRNAs have been reported to play potential roles in the dysfunction of endothelial cells and aSAH-related complications42,43,44,45,46. Recent studies suggested the absence of endothelial lining and the apoptosis of endothelial cells (ECs) in CVS18,47. Furthermore, ECs have been found to regulate the loss and erosion of cerebral arteries during vasospasm via micro RNAs (miRNAs)48,49. Abnormal miRNA expression levels have been implicated in various neurological diseases, including stroke, aneurysm, and vasospasm. Moreover, circulating miRNAs in blood have been shown to regulate inflammatory responses and affect cerebral arteries in CVS in patients with aSAH42,46,50. Considering the changes in their expression levels and their functional roles in blood, miRNAs represent promising candidates in the pathophysiology of CVS following aSAH. Therefore, we aim to explore miRNAs in whole-blood samples of aSAH patients during and outside of CVS, and investigate the molecular mechanisms of miRNAs in HBECs to gain a better understanding of CVS after aSAH.
Materials and methods
Patient selection and screening of cerebral vasospasm
This study was conducted in accordance with the ethical standards and approved by the Institutional Review Boards at the Brigham and Women’s Hospital (BWH) and informed consent was obtained from all patients. Six patients with aneurysmal subarachnoid hemorrhage whose maximal vasospasm was moderate to severe during their admission and who had sufficient samples for miRNA sequencing during and outside of vasospasm were included in the study. All patients received nimodipine until the resolution of vasospasm and underwent daily transcranial doppler ultrasonography (TCD). CT angiography (CTA) and digital subtraction angiography (DSA) were performed at least every other day during periods of moderate to severe vasospasm or as determined by the clinical team.
The following parameters were collected and analyzed: age, sex, race, smoking status, Hunt and Hess (H&H) scale, World Federal of Neurological Surgeon (WFNS) scale, and Fisher score (Table 1). Fisher score of 1–3 was used to assess for the amount of SAH without confounding with intraventricular hemorrhage.
Sample collection and experimental design
Blood samples were collected at various time points from six patients with aSAH at BWH between 2011 and 2016. Sample collection details are presented in Table S1. The degree of vasospasm for each sample was evaluated based on TCD values as previously described11. Briefly, in order to account for vasospasm in the entire Circle of Willis, rather than just the middle cerebral artery (MCA) alone, we categorized the samples using TCD and/or CTA results for MCA, anterior cerebral artery (ACA), posterior cerebral artery (PCA) that were done within 24 h of the blood samples. CTA was used when CTA was able to detect vasospasm in vessels not insonated by TCD. Samples obtained when TCDs had a mean flow velocity of MCA ≥ 150 cm/s, ACA ≥ 130 cm/s, or PCA ≥ 120 cm/s, namely samples with moderate to severe vasospasm, were categorized as CVS+. Samples with no or mild vasospasm, as well as some samples collected from patients at least one month after SAH were categorized as CVS−. CTA vasospasm was defined as vessels with > 25% narrowing compared to baseline admission CTA obtained before the onset of vasospasm.
MicroRNAs profiling
MicroRNAs (miRNAs) were isolated and purified from whole blood samples using the PAXgene Blood miRNA kit (Qiagen) according to the manufacturer’s standard protocol. MiRNA Libraries were prepared using the Bioo Scientific NEXTflex Small RNA-Seq kit v3 per the manufacturer’s instructions, and pooled libraries were sequenced on the Illumina HiSeq 2000 System to generate > 10 million 50 bp single end reads per sample, with a per sample target of ~ 2 million aligned miRNA reads. Raw data were subjected to quality control with FastQC (version 0.11.9)51, and to post-sequencing analysis using the bcbio-nextgen pipeline (version 1.2.9)52. The data in fastq format were aligned against human genome build GRCh38 using bowtie (version 1.3.1)53, and miRNA counts were estimated using seqcluster (version 1.2.8)54. MiRNA reads were annotated using the miRbase as the reference database for known miRNAs. MiRNAs without a count of 5 in at least 50% of samples were removed from further analysis.
Target prediction and miRNAs network analysis
Small RNA transcriptome profiles from CVS+ and CVS− blood samples were compared using R DESeq2 (version 1.36.0)55. MiRNAs with absolute Log2FoldChange > 1 and P < 0.05 were considered significant in this study. Over-representation analysis was conducted on these differentially expressed miRNAs (DEMis) using MIEAA2 (https://www.ccb.uni-saarland.de/mieaa2) with GO annotations derived from miRTarBase databases56. Terms with an adjusted P < 0.05 and an Expected Enrichment score ≥ 2 were considered significant. Targets of DEMis were obtained from miRTarBase by applying target cell type prediction using the TabularSapiens Celltype database and function prediction using the GO biological process database57,58,59,60. Results were visualized using R ggplot2 package61. Binding sites for miR-148b-3p on ROCK1 transcripts were identified using the RNA22 online server62. The parameters were adjusted to a sensitivity of 63% and a specificity of 61%. The seed region was set to 8 nt in length, with a tolerance for up to 2 unpaired bases within the seed. All other parameters were kept at their default settings.
Cell culture and transfection
Human brain endothelial cells (HBECs, ATCC) were cultured in DMEM: F-12 Medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco) at 37 °C with 5% CO2. HBECs were transfected with miR-NC mimic as a control or miR-148b-3p mimic (at final concentrations of 10, 20, and 50 nM, respectively) using lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s protocol. The cells were harvested 24 h after transfection. The sequences of the miRNA mimics used were as follows: miR-NC mimic (negative control), 5′-UCUACUUCUUUCUAGGACCUUGUCA-3′, and miR-148b-3p mimic, 5′-UCAGUGCAUCACAGAACUUUGU-3′.
Cell proliferation
Human brain endothelial cells (HBECs) were seeded at a density of 5 × 103 cells per well in a 96-well plate and treated with miR-NC mimic or miR-148b-3p mimic (20 nM) for 1, 2, 3, 4, and 5 days, respectively. The cells were evaluated for cell proliferation using a Water-Soluble Tetrazolium 1 (WST-1) assay (Roche) according to the manufacturer’s protocol. The absorbance was measured at 440 nm using a 96-well microplate reader (BioTeK Instruments).
Cell migration and invasion assays
Cell migration was evaluated using the wound healing assay. Human brain endothelial cells (HBECs) were seeded into a well with an insert to make a pre-casted wound, and then treated with miR-NC mimic or miR-148b-3p mimic (20 nM) for 24 h. The cells were cultured until a monolayer form, and then the inserts were removed to generate a wound field. The migration of cells into the wound field was monitored under a microscope. The wound area was measured and quantified using ImageJ63. To assess invasion ability, HBECs were seeded into trans-well insert (Corning) precoated with Matrigel (Corning). HBECs that invaded through the membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. The number of invading HBECs was measured under a microscope and quantified using ImageJ.
Real-time quantitative polymerase chain reaction
Total RNA, including miRNAs and mRNAs, was extracted using Trizol (Invirogen) according to the manufacturer’s protocol. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed to measure miRNA expression using the TaqMan miRNA reverse transcription kit according to the manufacturer’s protocol. MiRNA expression was normalized to U6. For mRNAs analysis, cDNA was amplified using SYBR Green Master Mix (Applied Biosystems) on the AB step one plus real-time PCR system (Applied Biosystems). mRNA expression was normalized to GAPDH. Each experiment was performed in triplicate. Relative miRNA and mRNA expression were calculated using the 2−ΔΔCq method. The primer sequences used were as follows: miRNA, reverse 5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTCAGG-3’; miR-148b-3p, forward 5’-GGGTCAGTGCATCACAGAA-3′; U6, forward 5′-TTCCTCCGCAAGGATGACACGC-3′; RhoA, forward 5′-AGCCTGTGGAAAGACATGCTT-3′ and reverse 5′-TCAAACACTGTGGGCACATAC-3′; ROCK1, forward 5′-AACATGCTGCTGGATAAATCTGG-3′ and reverse 5′-TGTATCACATCGTACCATGCCT-3′; GAPDH, forward 5′-CCCAGAATATCATCC-3′ and reverse 5′-CTGCTTCACCACCTTC-3′.
Western blot
Protein samples were prepared using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific), and protein concentration was measured using a BCA assay (Pierce). Total protein (20 µg per well) was separated by SDS-PAGE and transferred onto a PDVF membrane. After blocking with 5% BSA for 1 h, the membranes were incubated with primary antibodies: anti-RhoA, anti-ROCK1, anti-p-LIMK1, anti-LIMK, anti-p-Cofilin, and anti-Cofilin (Cell Signaling Technology). The membrane was visualized using ImageQuant LAS 500 (GE Healthcare).
Immunocytochemistry
Human brain endothelial cells (HBECs) were seeded onto glass coverslips, and treated with miR-NC mimic or miR-148b-3p mimic for 24 h. The cells were then fixed, permeabilized, and blocked. They were incubated with antibodies, including Alexa Fluor 488-conjugated phalloidin to visualize filamentous actin (F-actin) and Alexa Fluor 594-conjugated DNase I which selectively binds to globular actin (G-actin), overnight at 4 °C in the dark. Images were captured using a Leica TCS-SP5 confocal laser scanning microscope (CLSM).
Statistical analysis
Statistically significant differences in the results obtained from RT-qPCR analysis and the cell proliferation, migration and invasion assays were evaluated using an independent-samples t-test. The data were presented as the mean ± standard deviation (S.D.) from three independent experiments. P - values < 0.05 were considered to indicate a statistically significant difference unless otherwise stated. All calculations were performed using Prism (Graphpad Sotfware).
Results
Characteristics of aneurysmal subarachnoid hemorrhage (aSAH) patients and samples
The patient demographics are presented in Table 1. The mean age was 59.0 ± 15.8 years and all patients were female. The classification of aneurysmal subarachnoid hemorrhage (aSAH) severity at presentation was based on Hunt-Hess (H&H) scale, World Federal of Neurological Surgeon (WFNS) score, and Fisher score (Table 1). A total of 18 blood samples were collected from aSAH patients to perform miRNA profiling. We classified the samples into two groups relative to vasospasm status by transcranial doppler ultrasonography (TCD) and/or CT angiography (CTA) results (see Methods): eight samples were CVS− and ten samples were CVS+. The classification of samples revealed that CVS+ samples exhibited higher calculated Lindegaard (LR) ratio values and lower Glasgow Coma Scale (GCS) scores compared to CVS− samples (11.13 ± 3.14 vs. 10.38 ± 3.57). The CVS− samples were collected both during and outside the CVS time period (Table S1). The miRNA profiles showed no significant differences between these CVS− samples, regardless of whether they were collected during or outside the CVS time period (with adjusted P < 0.05 and Log2FoldChange > 2 as criteria for differential expression). The average Pearson’s correlation coefficient (PCC) between CVS− samples during and outside the CVS time period was 0.996 ± 0.004. Therefore, they were grouped together as one control group in this study.
Gene ontology (GO) enrichment analysis of differentially expressed miRNAs (DEMis)
To identify candidate miRNAs associated with CVS, we conducted miRNA sequencing analysis of CVS+ and CVS− samples. Differentially expressed miRNAs (DEMis) were screened and visualized using a volcano plot (Fig. 1a). We found three downregulated (miR-4732-3p, miR-16-2-3p, miR-6885-3p), and four upregulated (miR-29a-3p, miR-148b-3p, miR-374b-5p, miR-26b-5p) miRNAs in CVS+ compared to CVS− (P-value < 0.05). To further understand the biological functions of these DEMis in CVS, we performed over-representation analysis, which identified 62 gene ontology (GO) terms (Table S2). The resulting interaction network diagram displays 16 groups of GO terms (Fig. 1b), including response to stress, response to chemical, cytoskeleton organization, transport, RNA metabolic process, metabolic process, protein localization, cellular protein modification process, extracellular matrix organization, cell migration, vasculature development, signal transduction, biological process, cell proliferation and death, response to decreased oxygen levels, and platelet activation.
The expression profiles of miRNAs (a) The volcano plot displays significant differentially expressed miRNAs (DEMis) between CVS+ and CVS− samples. Red dots represent upregulated miRNAs, blue dots indicate downregulated miRNAs, and grey dots show no change. (b) Biological function network of DEMis with gene ontology (GO) terms as nodes linked. The node color depicts the significant enrichment. DEMis differentially expressed miRNAs, CVS+ blood samples with CVS, CVS− blood samples without CVS, GO gene ontology.
Prediction of potential target miR-148b-3p associated with CVS in DEMis
Recent studies have indicated that human brain endothelial cells (HBECs) lose their function in CVS following aSAH64. Moreover, it has been reported that cytoskeleton organization is essential for actin-based mobility of endothelial cells65,66. Figure 1b reveals that the target genes for CVS are associated with cell proliferation and death, cell migration, and cytoskeleton organization. Therefore, we hypothesized that there is a correlation between cell dysfunction and cytoskeleton organization that is regulated by the target DEMis. To explore this, we analyzed genes related to cellular cytoskeleton organization. The miRNA-target-enrichment network, which focuses on targets enriched in regulation of microtubule cytoskeleton organization, showed four upregulated miRNAs (miR-148b-3p, miR-374b-5p, miR-29a-3p, miR-26b-5p) and a downregulated miRNA (miR-16-2-3p) involved in this process (Fig. 2a). Among the target genes of these DEMis, ROCK1 is known to contribute to abnormal endothelial cells in cerebral and coronary vasospasm67,68, and to the regulation of vascular tone69,70. ROCK1 is also implicated in regulating phosphorylated LIMK within the Rho-ROCK pathway through phosphate metabolic processes. This regulary mechanism impacts actin cytoskeleton dynamics and cell migration in microvascular endothelial cells71. Thus, we further investigated the role of miR-148b-3p, the only DEMi targeting ROCK1.
Enriched biological processes of miR-148b-3p. (a) Gene network summarizes the miRNA targets belonging to the gene ontology (GO) term “regulation of microtubule cytoskeleton organization” as linked nodes. (b) Cell type enrichment analysis predicts the cell types regulated by miR-148b-3p. The bars indicate the transformed P-values of highly enriched cell types for each cell cluster (P-value < 0.05). (c) Chord diagram illustrates enriched biological processes of miR-148b-3p, linking each gene cluster to its corresponding processes. (d) miRNA expression levels of miR-148b-3p in patients’ samples using RT-qPCR. Error bars indicate mean ± S.D. of three independent experiments. miRNAs expressions were normalized to U6 expression. ***P < 0.001 when compared with CVS−. GO gene ontology, CVS+ blood samples with CVS, CVS− blood samples without CVS.
To explore which cell types miR-148b-3p mainly regulates, we conducted cell type enrichment analysis using miR-148b-3p targets (Fig. 2b). miR-148b-3p was predicted to regulate endothelial cells in various tissues. In addition, functional enrichment results showed that miR-148b-3p modulated as least 29 genes corresponding to processes involved in the negative regulation of smooth muscle cells (SMCs) proliferation, regulation of vascular associated SMCs proliferation, regulation of T cell mediated cytotoxicity, negative regulation of phosphate metabolic process, superoxide metabolic process, negative regulation of blood vessel endothelial cells (ECs) migration, negative regulation of abT cell differentiation, and pre-miRNA processing (Fig. 2c). In particular, miRNA-148b-3p presents a potential link between endothelial cell migration and associated genes. Given these observations, we focused our validation efforts on miR-148b-3p in patient samples. We confirmed miR-148b-3p expression levels in patients’ samples using real-time quantitative polymerase chain reaction (RT-qPCR) (Fig. 2d). MiR-148b-3p expression levels were increased in CVS+ compared to CVS−. These results showed that miR-148b-3p is highly expressed in aSAH during vasospasm, and may regulate ROCK1, thereby affecting cytoskeleton organization and migration in HBECs.
MiR-148b-3p suppresses cell growth, migration and invasion of HBECs
To investigate the role of miR-148b-3p in human brain endothelial cells (HBECs), we transfected HBECs with miRNA-148b-3p mimic or miRNA NC mimic (miR-NC) as a negative control to assess the biological effects of miRNA-148b-3p overexpression. We first verified the effects of transfection by RT-qPCR, which showed that miR-148b-3p mimic effectively altered miR-148-3p expression in HBECs using 20 nM and 50 nM concentrations for 24 h (Fig. S1). However, no significant difference was observed in the miR-NC group. Therefore, we used 20 nM of miR-148b-3p mimic for subsequent studies. Next, we evaluated the cell proliferation of HBECs over a 5-day period using a Water-Soluble Tetrazolium 1 (WST-1) assay (Fig. 3a). HBECs treated with the miR-148b-3p mimic exhibited suppressed cell growth compared to untreated cells and HBECs treated with the miR-NC. To assess the migration and invasion ability of miR-148b-3p affected HBECs, we conducted a wound healing assay and an invasion assay (Fig. 3b). The miR-148b-3p mimic-treated HBECs showed less wound closure compared to the miR-NC group. The closure percentage at the 24-h time point was significantly lower in the miR-148b-3p mimic group compared to untreated cells and miR-NC group (Fig. 3c). These results indicate that upregulation of miR-148b-3p led to a decrease in cell motility. Similarly, the number of HBECs penetrating through trans-well membranes was markedly reduced in the cells treated with miRNA-148-3p mimic compared to the other groups (Fig. 3d). The number of invading cells per field was approximately 2.1 times smaller in HBECs treated with the miR-148b-3p mimic than in the miR-NC group. These findings demonstrate that upregulation of miR-148b-3p significantly suppressed migration and invasion in HBECs.
MiR-148b-3p inhibits cell proliferation, migration, and invasion. (a) Effects of miR-148b-3p on cell proliferation in human brain endothelial cells (HBECs). The cell growth rates were determined for 5 days using a WST-1 assay in untreated HBECs (HBECs group), HBECs treated with miRNA-negative control mimic (miR-NC mi group), and HBECs treated with miRNA-148b-3p mimic (miR-148b-3p mi group). (b) Representative microscopic images of cell migration and invasion by miR-148b-3p in HBECs using a wound healing assay and a trans-well assay, respectively. (c) Percentage of the wound closure area at 24 h. (d) Quantification of the number of invasive cells per field. Error bars indicate mean ± S.D. of three independent experiments. **P < 0.01, ***P < 0.001 when compared with untreated HBECs or HBECs treated with miR-NC. miR-NC mi miRNA-negative control mimic, miR-148b-3p mi miRNA-148b-3p mimic.
MiR-148b-3p regulates ROCK-LIMK-Cofilin pathway in HBECs
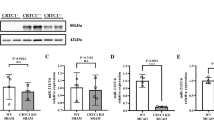
The ROCK-LIMK-Cofilin pathway is a well-known signaling pathway involved in the regulation of actin cytoskeleton, which plays a crucial role in cell motility and invasion72,73,74. Recent studies have suggested that miRNA-148b-3p directly targets rho-associated protein kinase 1 (ROCK1), and its upregulated expression is inversely associated with ROCK1 expression levels across multiple cell types75,76. miR-148b-3p is predicted to interact with the 3′UTR region of ROCK1 transcripts (Fig. S2). Additionally, our findings indicate the functional association between miR-148b-3p and ROCK1 in Fig. 2a,c. Therefore, we investigated whether miRNA-148b-3p affects the migration and invasion of HBECs through the ROCK-LIMK-Cofilin pathway. We found that the upregulation of miRNA-148b-3p did not affect the mRNA expression levels of RhoA, whereas the mRNA expression levels of ROCK were significantly decreased in HBECs treated with the miRNA-148b-3p mimic (Fig. 4a). We next confirmed the protein expression levels of RhoA, ROCK1, and other proteins associated with the ROCK-LIMK-Cofilin pathway by Western blot analysis (Fig. 4b,c and Fig. S3). The protein expression levels of RhoA, ROCK1 showed similar patterns to mRNA expression levels, with no difference observed in RhoA expression, while ROCK1 expression was decreased. We also assessed the total protein expression levels and the phosphorylation levels of LIM kinase 1 (LIMK1) and cofilin (Fig. 4b). ROCK1 directly phosphorylates LIMK1, and phosphorylated LIMK1 phosphorylates cofilin, which promotes cell motility77. Therefore, we quantified the phosphorylation levels using the total protein and phosphorylated protein by ImageJ. The phosphorylation of LIMK1 and cofilin were decreased by the miRNA-148b-3p (Fig. 4c). Our results demonstrate that miRNA-148b-3p targets ROCK1 and decreases its expression, which leads to a decrease in the phosphorylation of LIMK1 and cofilin. We then investigated the change in globular actin (G-actin) and fibrous actin (F-actin) in HBECs using confocal laser scanning microscopy (Fig. 4d). In HBEC treated with miR-148b-3p mimic, the G-actin levels increased, and the F-actin exhibited abnormalities in the filament structure. Notably, the morphology of HBECs treated with miR-148b-3p mimic appeared smaller and more contracted compared to the control cells. Overall, our study showed that miR-148b-3p can suppress cell growth, migration, and invasion of HBECs by inhibiting the expression of the ROCK-LIMK-Cofilin pathway and disrupting the reorganization of actin cytoskeleton (Fig. 4e).
MiR-148b-3p affects ROCK-LIMK-Cofilin pathway in human brain endothelial cells (HBECs). (a) mRNA expression levels of RhoA and ROCK1 in untreated HBECs, and HBECs treated with miR-NC or miR-148b-3p mimic using RT-qPCR. (b) Representative western blot images of protein expression of markers associated with ROCK-LIMK-Cofilin pathway. (c) Quantification of protein expression levels using ImageJ. (d) Representative confocal images of untreated HBECs, and HBECs treated with miR-NC or miR-148b-3p mimic after immunostaining for G-actin (red), F-actin (green), and nuclei (blue). (e) Schematic diagram of this study. Overall, we discovered that miRNA-148b-3p is highly expressed in aSAH patients during vasospasm, and that it regulates cell proliferation, migration, and invasion in human brain endothelial cells. Error bars indicate mean ± S.D. of three independent experiments. mRNAs expressions were normalized to GAPDH expression. **P < 0.01, ***P < 0.001 when compared with untreated HBECs or HBECs treated with miR-NC. miR-NC mi miRNA-negative control mimic, miR-148b-3p mi miRNA-148b-3p mimic, ROCK Rho-associated protein kinase, LIMK LIM kinase, G-actin globular actin, F-actin fibrous actin, DAPI 4’,6-diamidino-2-phenylindole.
Discussion
Given that several miRNAs influence endothelial dysfunction and complications in aSAH, including changes in miRNA expression associated with CVS, we aim to explore miRNAs in whole-blood samples and HBECs to better understand their roles in CVS following aSAH. We performed miRNA profiling in whole-blood samples from aSAH patients whose maximal vasospasms were moderate or severe. MiRNA profiles of blood samples obtained during and outside of vasospasm were compared. The analysis revealed several miRNAs with differential expression between the two groups. Notably, miR-148b-3p, the only DEMi that targets rho-associated protein kinase 1 (ROCK1) among the target genes of DEMis, affects endothelial and vascular tone regulation in vasospasm, and was identified as an upregulated miRNA in patients during vasospasm. Moreover, miR-148b-3p was predicted to target endothelial cells in various organs and play a role in cytoskeleton organization.
Recent studies indicate that HBECs dysfunction in CVS following aSAH, and that cytoskeleton organization is crucial for actin-based mobility of endothelial cells78,79. Therefore, we further investigated the potential role of miR-148b-3p in human brain endothelial cells (HBECs). Our findings demonstrated that upregulation of miR-148b-3p resulted in the suppression of cell growth, migration, and invasion in HBECs, indicating its potential involvement in vasospasm. To gain a deeper understanding of the molecular mechanisms underlying the effects of miR-148b-3p on HBECs, we focused on the ROCK-LIMK-Cofilin pathway, which plays a critical role in cytoskeletal organization and cell motility. Our results indicated that miR-148b-3p directly targeted and decreased the expression of ROCK1. The downregulation of ROCK1 led to a reduction in the phosphorylation of downstream targets LIM kinase 1 (LIMK1) and cofilin. ROCK1 is known to regulate permeability of endothelial cells, and contribute to abnormal endothelial cells in cerebral vasculature67,80,81. Some studies have demonstrated that endothelial damage in the major cerebral arteries is associated with the pathogenesis of vasospasm82. The disruption of the ROCK-LIMK-Cofilin pathway results in abnormal actin cytoskeletal organization, ultimately leading to dysfunction of ECs, which can contribute to the occurrence of vasospasm67,80,81,82. Overall, our study highlights the involvement of miR-148b-3p in the regulation of HBECs function, enhancing our understanding of its potential role in the development of vasospasm.
Interestingly, cofilin has been found to be involved other types of brain injury, including intracerebral hemorrhage and traumatic brain injury, by affecting inflammation and synaptic plasticity. Cofilin has been found to mediate neuroinflammation and affect synaptic loss in intracerebral hemorrhage where a cofilin inhibitor has been found to improve neurological outcomes83. Cofilin has also been found to be involved in synaptic plasticity via NMDA receptor-cofilin signaling84 and via phosphatase calcineurin-cofilin signaling85 in traumatic brain injury. Thus while this study mainly elucidates the role of the ROCK-LIMK-Cofilin pathway in EC dysfunction, the effects may involve neuronal plasticity and inflammation.
This study’s main limitation is the small number of participants. Thus, these findings require validation in larger patient cohorts. However, we had restricted our patient cohort to the more extreme phenotypes, specifically patients whose peak vasospasms were moderate or severe, to maximize the yield of the analyses. All patients included in this study were female due to the high predominance of female patients with aneurysmal SAH. Female patients with aSAH have a worse prognosis compared to male patients, which may be due to more susceptibility to cerebral ischemia57,86,87. Given the limited sample size of this study, having an all female cohort would minimize the potential confounding effects related to sex differences. However, future research is required to understand sex differences in the development of complications associated with CVS. We also ensured the accuracy of miRNA sequencing and implemented thresholds for filtering low-quality data. In addition, we focused our attention on miR-148b-3p in HBECs. However, it is important to consider that other miRNAs and environmental factors, such as hypoxia88,89,90, may also have significant effects. Additionally, contributions from the other major vascular cell types, such as smooth muscle cells and fibroblasts91,92,93, need to be further studied. In vivo studies are necessary to evaluate the potential of miR-148b-3p and its associated pathway as potential therapeutic targets. Finally, this study focused on cerebral vasospasm. Although vasospasm is a major contributor to DCI, there are a number of other potential contributors such as microthrombosis, cortical spreading depression, neuroinflammation and microcirculatory dysfunction3. Future studies will be needed to examine all aspects of DCI.
Our study revealed the upregulation of miR-148b-3p in aSAH patients during vasospasm. We demonstrated that miR-148b-3p regulates the ROCK-LIMK-Cofilin pathway, which influences the proliferation, migration, and invasion of HBECs. These findings suggest the potential of miR-148b-3p as a diagnostic marker and therapeutic target for addressing endothelial cells dysfunction in vasospasm following aSAH.
Data availability
MiRNA sequencing data are available at SRA (weblink: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1095482?reviewer=grv12u712mq53df55f6577rvta). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
de Rooij, N. K., Linn, F. H., van der Plas, J. A., Algra, A. & Rinkel, G. J. Incidence of subarachnoid haemorrhage: a systematic review with emphasis on region, age, gender and time trends. J. Neurol. Neurosurg. Psychiatry. 78, 1365–1372. https://doi.org/10.1136/jnnp.2007.117655 (2007).
Foreman, B. The pathophysiology of delayed cerebral ischemia. J. Clin. Neurophysiol. 33, 174–182. https://doi.org/10.1097/WNP.0000000000000273 (2016).
Geraghty, J. R. & Testai, F. D. Delayed cerebral ischemia after subarachnoid hemorrhage: beyond vasospasm and towards a multifactorial pathophysiology. Curr. Atheroscler Rep. 19, 50. https://doi.org/10.1007/s11883-017-0690-x (2017).
Kassell, N. F. et al. The International Cooperative Study on the timing of aneurysm surgery. Part 1: overall management results. J. Neurosurg. 73, 18–36. https://doi.org/10.3171/jns.1990.73.1.0018 (1990).
Dorsch, N. W. & King, M. T. A review of cerebral vasospasm in aneurysmal subarachnoid haemorrhage part I: incidence and effects. J. Clin. Neurosci. 1, 19–26. https://doi.org/10.1016/0967-5868(94)90005-1 (1994).
Rabinstein, A. A. et al. Predictors of cerebral infarction in aneurysmal subarachnoid hemorrhage. Stroke. 35, 1862–1866. https://doi.org/10.1161/01.STR.0000133132.76983.8e (2004).
Macdonald, R. L., Pluta, R. M. & Zhang, J. H. Cerebral vasospasm after subarachnoid hemorrhage: the emerging revolution. Nat. Clin. Pract. Neurol. 3, 256–263. https://doi.org/10.1038/ncpneuro0490 (2007).
Pluta, R. M. et al. Cerebral vasospasm following subarachnoid hemorrhage: time for a new world of thought. Neurol. Res. 31, 151–158. https://doi.org/10.1179/174313209X393564 (2009).
Vajkoczy, P., Horn, P., Thome, C., Munch, E. & Schmiedek, P. Regional cerebral blood flow monitoring in the diagnosis of delayed ischemia following aneurysmal subarachnoid hemorrhage. J. Neurosurg. 98, 1227–1234. https://doi.org/10.3171/jns.2003.98.6.1227 (2003).
Castanares-Zapatero, D. & Hantson, P. Pharmacological treatment of delayed cerebral ischemia and vasospasm in subarachnoid hemorrhage. Ann. Intensive Care. 1, 12. https://doi.org/10.1186/2110-5820-1-12 (2011).
Snider, S. B. et al. Transcranial-doppler-measured vasospasm severity is Associated with delayed cerebral infarction after subarachnoid hemorrhage. Neurocrit Care. 36, 815–821. https://doi.org/10.1007/s12028-021-01382-2 (2022).
Yoshimoto, Y., Tanaka, Y. & Hoya, K. Acute systemic inflammatory response syndrome in subarachnoid hemorrhage. Stroke. 32, 1989–1993. https://doi.org/10.1161/hs0901.095646 (2001).
Dhar, R. & Diringer, M. N. The burden of the systemic inflammatory response predicts vasospasm and outcome after subarachnoid hemorrhage. Neurocrit Care. 8, 404–412. https://doi.org/10.1007/s12028-008-9054-2 (2008).
Diringer, M. N. Subarachnoid hemorrhage: a multiple-organ system disease. Crit. Care Med. 31, 1884–1885. https://doi.org/10.1097/01.CCM.0000063528.09569.3A (2003).
Ji, R. et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 100, 1579–1588. https://doi.org/10.1161/CIRCRESAHA.106.141986 (2007).
Small, E. M. & Olson, E. N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 469, 336–342. https://doi.org/10.1038/nature09783 (2011).
Mayberg, M. R., Okada, T. & Bark, D. H. The significance of morphological changes in cerebral arteries after subarachnoid hemorrhage. J. Neurosurg. 72, 626–633. https://doi.org/10.3171/jns.1990.72.4.0626 (1990).
Zubkov, A. Y., Ogihara, K., Bernanke, D. H., Parent, A. D. & Zhang, J. Apoptosis of endothelial cells in vessels affected by cerebral vasospasm. Surg. Neurol. 53, 260–266. https://doi.org/10.1016/s0090-3019(99)00187-1 (2000).
Schneider, U. C., Xu, R. & Vajkoczy, P. Inflammatory events following subarachnoid hemorrhage (SAH). Curr. Neuropharmacol. 16, 1385–1395. https://doi.org/10.2174/1570159X16666180412110919 (2018).
Choi, S. J. & Lillicrap, D. A sticky proposition: the endothelial glycocalyx and von Willebrand factor. J. Thromb. Haemost. 18, 781–785. https://doi.org/10.1111/jth.14743 (2020).
Ishikawa, M. et al. Platelet and leukocyte adhesion in the microvasculature at the cerebral surface immediately after subarachnoid hemorrhage. Neurosurgery. 64, 546–553. https://doi.org/10.1227/01.NEU.0000337579.05110.F4 (2009). discussion 553 – 544.
Sanicola, H. W. et al. Pathophysiology, management, and therapeutics in subarachnoid hemorrhage and delayed cerebral ischemia: an overview. Pathophysiology. 30, 420–442. https://doi.org/10.3390/pathophysiology30030032 (2023).
Suzuki, H. et al. Cerebrovascular pathophysiology of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage. Histol. Histopathol. 36, 143–158. https://doi.org/10.14670/HH-18-253 (2021).
Schenck, H. et al. The role of the glycocalyx in the pathophysiology of Subarachnoid Hemorrhage-Induced delayed cerebral ischemia. Front. Cell. Dev. Biol. 9, 731641. https://doi.org/10.3389/fcell.2021.731641 (2021).
Iorio, M. V. et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707. https://doi.org/10.1158/0008-5472.CAN-07-1936 (2007).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell. 136, 215–233. https://doi.org/10.1016/j.cell.2009.01.002 (2009).
Chen, R., Wang, G., Zheng, Y., Hua, Y. & Cai, Z. Long non-coding RNAs in osteosarcoma. Oncotarget. 8, 20462–20475. https://doi.org/10.18632/oncotarget.14726 (2017).
Kin, K. et al. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J. Am. Heart Assoc. 1, e000745. https://doi.org/10.1161/JAHA.112.000745 (2012).
Santoro, M. M. & Nicoli, S. miRNAs in endothelial cell signaling: the endomiRNAs. Exp. Cell. Res. 319, 1324–1330. https://doi.org/10.1016/j.yexcr.2012.12.009 (2013).
Tan, J. R. et al. microRNAs in stroke pathogenesis. Curr. Mol. Med. 11, 76–92. https://doi.org/10.2174/156652411794859232 (2011).
Koutsis, G., Siasos, G. & Spengos, K. The emerging role of microRNA in stroke. Curr. Top. Med. Chem. 13, 1573–1588. https://doi.org/10.2174/15680266113139990106 (2013).
Li, H. T. et al. Upregulation of microRNA–24 causes vasospasm following subarachnoid hemorrhage by suppressing the expression of endothelial nitric oxide synthase. Mol. Med. Rep. 18, 1181–1187. https://doi.org/10.3892/mmr.2018.9050 (2018).
Liu, D. et al. Genome-wide microRNA changes in human intracranial aneurysms. BMC Neurol. 14, 188. https://doi.org/10.1186/s12883-014-0188-x (2014).
Vikman, P., Beg, S., Khurana, T. S., Hansen-Schwartz, J. & Edvinsson, L. Gene expression and molecular changes in cerebral arteries following subarachnoid hemorrhage in the rat. J. Neurosurg. 105, 438–444. https://doi.org/10.3171/jns.2006.105.3.438 (2006).
Miller, B. A., Turan, N., Chau, M. & Pradilla, G. Inflammation, vasospasm, and brain injury after subarachnoid hemorrhage. Biomed. Res. Int. 2014(384342), 1. https://doi.org/10.1155/2014/384342 (2014).
Khoshnam, S. E., Winlow, W., Farbood, Y., Moghaddam, H. F. & Farzaneh, M. Emerging roles of microRNAs in ischemic stroke: as possible therapeutic agents. J. Stroke. 19, 166–187. https://doi.org/10.5853/jos.2016.01368 (2017).
Lopes, K. P. et al. Global miRNA expression profile reveals novel molecular players in aneurysmal subarachnoid haemorrhage. Sci. Rep. 8, 8786. https://doi.org/10.1038/s41598-018-27078-w (2018).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5 (2004).
Fichtlscherer, S., Zeiher, A. M. & Dimmeler, S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb. Vasc Biol. 31, 2383–2390. https://doi.org/10.1161/ATVBAHA.111.226696 (2011).
Creemers, E. E., Tijsen, A. J. & Pinto, Y. M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 110, 483–495. https://doi.org/10.1161/CIRCRESAHA.111.247452 (2012).
Zampetaki, A., Willeit, P., Drozdov, I., Kiechl, S. & Mayr, M. Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc. Res. 93, 555–562. https://doi.org/10.1093/cvr/cvr266 (2012).
Su, X. W. et al. Circulating microRNA 132-3p and 324-3p profiles in patients after Acute Aneurysmal Subarachnoid Hemorrhage. PLoS One. 10, e0144724. https://doi.org/10.1371/journal.pone.0144724 (2015).
Powers, C. J. et al. Human cerebrospinal fluid microRNA: temporal changes following subarachnoid hemorrhage. Physiol. Genomics. 48, 361–366. https://doi.org/10.1152/physiolgenomics.00052.2015 (2016).
Bache, S. et al. MicroRNA changes in cerebrospinal fluid after subarachnoid hemorrhage. Stroke 48, 2391–2398. https://doi.org/10.1161/STROKEAHA.117.017804 (2017).
Pulcrano-Nicolas, A. S. et al. Whole-blood miRNA sequencing profiling for vasospasm in patients with aneurysmal subarachnoid hemorrhage. Stroke 49, 2220–2223. https://doi.org/10.1161/STROKEAHA.118.021101 (2018).
Sheng, B. et al. Early serum miR-1297 is an indicator of poor neurological outcome in patients with aSAH. Biosci. Rep. 38, 1. https://doi.org/10.1042/BSR20180646 (2018).
Zhou, C. et al. Caspase inhibitors prevent endothelial apoptosis and cerebral vasospasm in dog model of experimental subarachnoid hemorrhage. J. Cereb. Blood Flow. Metab. 24, 419–431. https://doi.org/10.1097/00004647-200404000-00007 (2004).
Sun, L. et al. The expression of cerebrospinal fluid exosomal miR-630 plays an important role in the dysfunction of endothelial cells after subarachnoid hemorrhage. Sci. Rep. 9, 11510. https://doi.org/10.1038/s41598-019-48049-9 (2019).
Frijns, C. J. et al. Endothelial cell activation after subarachnoid hemorrhage. Neurosurgery 50, 1223–1229 (discussion 1229–1230). https://doi.org/10.1097/00006123-200206000-00009 (2002).
Stylli, S. S. et al. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J. Neurosurg. 126, 1131–1139. https://doi.org/10.3171/2016.1.JNS151454 (2017).
Andrews, S. & FastQC A quality control tool for high throughput sequence data. citeulike-article-id:11583827
Chapman, B. et al. bcbio/bcbio-nextgen: (v1.2.9). Zenodo. https://doi.org/10.5281/zenodo.5781867 (2021).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Pantano, L., Estivill, X. & Marti, E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics. 27, 3202–3203. https://doi.org/10.1093/bioinformatics/btr527 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Aparicio-Puerta, E. et al. miEAA 2023: updates, new functional microRNA sets and improved enrichment visualizations. Nucleic Acids Res. 51, W319–W325. https://doi.org/10.1093/nar/gkad392 (2023).
Huang, H. Y. et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 50, D222–D230. https://doi.org/10.1093/nar/gkab1079 (2022).
Tabula Sapiens, C. et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 376, eabl4896. https://doi.org/10.1126/science.abl4896 (2022).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Gene Ontology, C. et al. The Gene Ontology knowledgebase in 2023. Genetics. 224. https://doi.org/10.1093/genetics/iyad031 (2023).
Wickham, H. Ggplot2: Elegant Graphics for data Analysis (Springer, 2016).
Miranda, K. C. et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 126, 1203–1217. https://doi.org/10.1016/j.cell.2006.07.031 (2006).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675. https://doi.org/10.1038/nmeth.2089 (2012).
Clower, B. R., Yamamoto, Y., Cain, L., Haines, D. E. & Smith, R. R. Endothelial injury following experimental subarachnoid hemorrhage in rats: effects on brain blood flow. Anat. Rec. 240, 104–114. https://doi.org/10.1002/ar.1092400110 (1994).
Rajendran, P. et al. The vascular endothelium and human diseases. Int. J. Biol. Sci. 9, 1057–1069. https://doi.org/10.7150/ijbs.7502 (2013).
Saner, H., Wurbel, H., Mahler, F., Flammer, J. & Gasser, P. Microvasculatory evaluation of vasospastic syndromes. Adv. Exp. Med. Biol. 220, 215–218. https://doi.org/10.1007/978-1-4613-1927-6_38 (1987).
Sato, M., Tani, E., Fujikawa, H. & Kaibuchi, K. Involvement of rho-kinase-mediated phosphorylation of myosin light chain in enhancement of cerebral vasospasm. Circ. Res. 87, 195–200. https://doi.org/10.1161/01.res.87.3.195 (2000).
Liao, J. K., Seto, M. & Noma, K. Rho kinase (ROCK) inhibitors. J. Cardiovasc. Pharmacol. 50, 17–24. https://doi.org/10.1097/FJC.0b013e318070d1bd (2007).
Takemoto, M., Sun, J., Hiroki, J., Shimokawa, H. & Liao, J. K. Rho-kinase mediates hypoxia-induced downregulation of endothelial nitric oxide synthase. Circulation. 106, 57–62. https://doi.org/10.1161/01.cir.0000020682.73694.ab (2002).
Rikitake, Y. et al. Inhibition of rho kinase (ROCK) leads to increased cerebral blood flow and stroke protection. Stroke. 36, 2251–2257. https://doi.org/10.1161/01.STR.0000181077.84981.11 (2005).
Dvorsky, R., Blumenstein, L., Vetter, I. R. & Ahmadian, M. R. Structural insights into the interaction of ROCKI with the switch regions of RhoA. J. Biol. Chem. 279, 7098–7104. https://doi.org/10.1074/jbc.M311911200 (2004).
Itoh, K. et al. An essential part for rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 5, 221–225. https://doi.org/10.1038/5587 (1999).
Olson, M. F. & Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis. 26, 273–287. https://doi.org/10.1007/s10585-008-9174-2 (2009).
Kumper, S. et al. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. Elife. 5, e12994. https://doi.org/10.7554/eLife.12203 (2016).
Chen, X., Bo, L., Lu, W., Zhou, G. & Chen, Q. MicroRNA-148b targets rho-associated protein kinase 1 to inhibit cell proliferation, migration and invasion in hepatocellular carcinoma. Mol. Med. Rep. 13, 477–482. https://doi.org/10.3892/mmr.2015.4500 (2016).
Dorraki, N. et al. miRNA-148b and its role in various cancers. Epigenomics. 13, 1939–1960. https://doi.org/10.2217/epi-2021-0155 (2021).
Bessueille, L. & Magne, D. Inflammation: a culprit for vascular calcification in atherosclerosis and diabetes. Cell. Mol. Life Sci. 72, 2475–2489. https://doi.org/10.1007/s00018-015-1876-4 (2015).
Iuliano, B. A., Pluta, R. M., Jung, C. & Oldfield, E. H. Endothelial dysfunction in a primate model of cerebral vasospasm. J. Neurosurg. 100, 287–294. https://doi.org/10.3171/jns.2004.100.2.0287 (2004).
Viderman, D., Tapinova, K. & Abdildin, Y. G. Mechanisms of cerebral vasospasm and cerebral ischaemia in subarachnoid haemorrhage. Clin. Physiol. Funct. Imaging. 43, 1–9. https://doi.org/10.1111/cpf.12787 (2023).
Wojciak-Stothard, B. & Ridley, A. J. Rho GTPases and the regulation of endothelial permeability. Vascul Pharmacol. 39, 187–199. https://doi.org/10.1016/s1537-1891(03)00008-9 (2002).
Noma, K., Oyama, N. & Liao, J. K. Physiological role of ROCKs in the cardiovascular system. Am. J. Physiol. Cell. Physiol. 290, C661–668. https://doi.org/10.1152/ajpcell.00459.2005 (2006).
Sasaki, T. & Kassell, N. F. The role of endothelium in cerebral vasospasm. Neurosurg. Clin. N Am. 1, 451–463 (1990).
Almarghalani, D. A., Bahader, G. A., Ali, M., Tillekeratne, L. M. V. & Shah, Z. A. Cofilin inhibitor improves neurological and cognitive functions after Intracerebral Hemorrhage by suppressing endoplasmic reticulum stress related-neuroinflammation. Pharmaceuticals (Basel). 17. https://doi.org/10.3390/ph17010114 (2024).
Smith, M. et al. Blast waves from detonated military explosive reduce GluR1 and synaptophysin levels in hippocampal slice cultures. Exp. Neurol. 286, 107–115. https://doi.org/10.1016/j.expneurol.2016.10.002 (2016).
Campbell, J. N. et al. Mechanisms of dendritic spine remodeling in a rat model of traumatic brain injury. J. Neurotrauma. 29, 218–234. https://doi.org/10.1089/neu.2011.1762 (2012).
Kongable, G. L. et al. Gender-related differences in aneurysmal subarachnoid hemorrhage. J. Neurosurg. 84, 43–48. https://doi.org/10.3171/jns.1996.84.1.0043 (1996).
Cai, Y. et al. Comparison of sex differences in outcomes of patients with Aneurysmal Subarachnoid Hemorrhage: a single-Center Retrospective Study. Front. Neurol. 13, 853513. https://doi.org/10.3389/fneur.2022.853513 (2022).
Yan, J. et al. 2-methoxyestradiol reduces cerebral vasospasm after 48 hours of experimental subarachnoid hemorrhage in rats. Exp. Neurol. 202, 348–356. https://doi.org/10.1016/j.expneurol.2006.06.009 (2006).
Vellimana, A. K. et al. Endothelial nitric oxide synthase mediates endogenous protection against subarachnoid hemorrhage-induced cerebral vasospasm. Stroke. 42, 776–782. https://doi.org/10.1161/STROKEAHA.110.607200 (2011).
Milner, E. et al. HIF-1alpha mediates isoflurane-Induced Vascular Protection in Subarachnoid Hemorrhage. Ann. Clin. Transl Neurol. 2, 325–337. https://doi.org/10.1002/acn3.170 (2015).
Cipolla, M. J. & Osol, G. Vascular smooth muscle actin cytoskeleton in cerebral artery forced dilatation. Stroke. 29, 1223–1228. https://doi.org/10.1161/01.str.29.6.1223 (1998).
Walsh, M. P. & Cole, W. C. The role of actin filament dynamics in the myogenic response of cerebral resistance arteries. J. Cereb. Blood Flow. Metab. 33, 1–12. https://doi.org/10.1038/jcbfm.2012.144 (2013).
Saphirstein, R. J., Gao, Y. Z., Lin, Q. Q. & Morgan, K. G. Cortical actin regulation modulates vascular contractility and compliance in veins. J. Physiol. 593, 3929–3941. https://doi.org/10.1113/JP270845 (2015).
Acknowledgements
Blood samples were obtained in conjunction with Partners Healthcare Biobank.
Funding
This study was supported by a grant from Partners Personalized Medicine (RD).
Author information
Authors and Affiliations
Contributions
JR, JZ and RD wrote and revised the manuscript and contributed to study design, acquisition of data, analysis of data. ST, SD, KF, NP, MA, AB, MR, SA and SW provide meaningful input to the manuscript and contributed to analysis and acquisition and interpretation of data. All authors reviewed and contributed to manuscript drafts and revisions.
Corresponding author
Ethics declarations
Competing interests
MAA: Proctor for Covidien and Codman. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ryu, JY., Zhang, J., Tirado, SR. et al. MiRNA expression profiling reveals a potential role of microRNA-148b-3p in cerebral vasospasm in subarachnoid hemorrhage. Sci Rep 14, 22539 (2024). https://doi.org/10.1038/s41598-024-73579-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73579-2