Abstract
Recognition of viral infection often relies on the detection of double-stranded RNA (dsRNA), a process that is conserved in many different organisms. In mammals, proteins such as MDA5, RIG-I, OAS, and PKR detect viral dsRNA, but struggle to differentiate between viral and endogenous dsRNA. This study investigates an shRNA targeting DDX54’s potential to activate PKR, a key player in the immune response to dsRNA. Knockdown of DDX54 by a specific shRNA induced robust PKR activation in human cells, even when DDX54 is overexpressed, suggesting an off-target mechanism. Activation of PKR by the shRNA was enhanced by knockdown of ADAR1, a dsRNA binding protein that suppresses PKR activation, indicating a dsRNA-mediated mechanism. In vitro assays confirmed direct PKR activation by the shRNA. These findings emphasize the need for rigorous controls and alternative methods to validate gene function and minimize unintended immune pathway activation.
Similar content being viewed by others
Introduction
Recognition and response to viral infection are essential processes in all kingdoms of life. For organisms infected by viruses, recognition of viral infection is often dependent upon sensing double stranded RNAs (dsRNA) generated from the viral genome. In mammals, multiple sensor proteins detect viral dsRNA. The RIG-I like receptors MDA5 and RIG-I, activate the type I interferon (IFN) pathway following detection of viral dsRNA1. Oligoadenylate synthase (OAS) proteins activate RNase L2, while protein kinase R (PKR) activates the integrated stress response pathway after sensing viral dsRNA3. Each of these pathways relies on a sensor protein binding to dsRNA; however, these proteins cannot distinguish between viral dsRNA (exogenous) and dsRNAs arising from inter- and intramolecular base-pairing between endogenous RNAs4.
Endogenous dsRNAs arise from a variety of RNA transcripts. While some endogenous dsRNAs are formed through base-pairing interactions between repetitive elements, such as inverted Alu repeats5, other, often much shorter, endogenous dsRNAs are intermediates of the microRNA (miRNA) biogenesis pathway6. Primary miRNA (pri-miRNA) transcripts are generally transcribed by RNA polymerase II and form stem-loop structures with mismatches, loops and bulges. These pri-miRNAs are processed by DROSHA and its partner protein DGCR8 in the nucleus to form stem-loop structures of between ~ 55–70 nucleotides long, referred to as precursor miRNAs (pre-miRNA). Finally, the pre-miRNA is processed further in the cytoplasm by DICER and its accessory proteins to form mature miRNAs that are loaded on to an argonaute protein to form the miRNA-induced silencing complex (miRISC). The components of miRNA processing, specifically DICER, also processes exogenous short-hairpin RNAs (shRNAs)7. By using the cellular machinery associated with miRNA processing and miRNA-mediated repression, shRNAs are routinely used to knockdown the expression of genes of interest. While shRNAs can provide robust knockdown of target genes, they have been plagued by off-target effects, usually through miRNA like effects on non-target mRNAs8,9,10.
The protein double-stranded RNA dependent protein kinase (PKR) serves an important role in sensing viral dsRNA. PKR is the primary mechanism of the innate immune system to trigger apoptosis and inhibit protein synthesis11,12,13. PKR contains three domains: two dsRNA binding domains (dsRBD), dsRBD1 and dsRBD2, and a kinase domain14. Activation of PKR requires dsRNA binding, dimerization, and autophosphorylation15. Upon binding of dsRNA by two PKR monomers, PKR dimerizes, bringing the kinase domains of each monomer close enough to autophosphorylate and activate PKR16,17. Active PKR has several canonical substrates, including the alpha subunit of eukaryotic initiation factor 2 (eIF2α), which when phosphorylated by PKR inhibits translation18. Key to PKR activation are its dsRBDs. Each dsRBD is capable of binding dsRNA in a sequence independent manner, and can bind dsRNAs as short as 16 bp19,20. While PKR has an important role in sensing viral dsRNAs, because the dsRBDs bind in a sequence-independent manner PKR can also bind numerous endogenous dsRNAs21. Here, we provide evidence for a shRNA that directly activates PKR in human cell lines.
Results
Knockdown of DDX54 causes activation of PKR
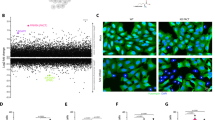
Using proximity labeling, we had previously identified the RNA helicase DDX54 as an ADAR1 interacting protein22. This led us to study the role of DDX54 in the regulation of dsRNA sensing, a role that has been well established for ADAR1. To begin to evaluate the role of DDX54, we utilized lentiviral shRNA mediated knockdown by RNAi to reduce DDX54 expression in two ADAR1-dependent cell lines. These cells require ADAR1 for viability and will activate multiple dsRNA sensing pathways following genetic depletion of ADAR123. One specific shRNA sequence showed substantial knockdown of DDX54 expression, Fig. 1a. This shRNA sequence is encoded within two shRNA expression plasmids from The RNAi Consortium (TRC) library, TRCN0000049957 and TRCN0000289044, referred to here as shDDX54-4 and shDDX54-5; although the plasmids encoding these shRNAs are slightly different, the shRNA itself is identical in sequence. Interestingly, we observed robust activation of PKR, as indicated by PKR phosphorylation at Thr446 and phosphorylation of eIF2α at Ser51, in cells transduced with lentivirus encoding shDDX54-4 and shDDX54-5, Fig. 1a,b. In addition to activation of PKR, we observed reduced cellular proliferation, and/or viability, as indicated by reduced foci formation, Fig. 1c,d. In BT549 we also observed increased cleaved PARP (c-PARP, 89 kDa protein produced by cleavage after Asp214 by caspase24,25), indicating those cells are dying from apoptosis. These observations were consistent with the effect of ADAR1 knockdown in these same cell lines23.
An shRNA targeting DDX54 causes activation of PKR. (a) Representative immunoblot of pPKR and other proteins of interest following knockdown of DDX54 by shDDX54-4 and shDDX54-5 (identical shRNA sequence) in MDA-MB-231 and BT549. Total protein was imaged using a Stain-Free Gel and was used for normalization. (b) Quantification of pPKR/PKR from the immunoblot in (a). See Supplemental Fig. 1 for quantification of other proteins of interest. (c) Quantification of foci formation shown in (d). (e) Representative immunoblot of pPKR and other proteins of interest following knockdown of DDX54 by shDDX54-4 in empty vector (EV) control MDA-MB-231 or DDX54 overexpressing (DDX54-OE) MDA-MB-231. (f) Cell proliferation as measured by counting viable cells by trypan blue exclusion. (g) Quantification of immunoblot in (e). (h) Representative immunoblot of pPKR and other proteins of interest following knockdown of DDX54 by shDDX54-4 in empty vector (EV) control BT549 or DDX54 overexpressing (DDX54-OE) BT549. (i) Quantification of immunoblot in (h). Bars or larger points represent the average of at least three replicates (shown as differently shaped points), error bars are ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. P-values determined by Dunnett’s test (b,c) or one-way ANOVA with post-hoc Tukey (g–i).
PKR activation is an off-target effect of the shRNA targeting DDX54
Based on the DDX54 knockdown phenotype, we began to explore the mechanism for PKR activation following knockdown of DDX54 in BT549. To verify that the effects we had observed were caused by reduced DDX54 expression, and to rule out off-target effects of the shRNA, we performed a rescue experiment. We overexpressed a wobble mutant of DDX54 (not targetable by shDDX54-4) and knocked down endogenous DDX54 using shDDX54-4 in both MDA-MB-231 and BT549. Surprisingly, we observed that exogenous DDX54 expression was not sufficient to prevent activation of PKR in cells expressing shDDX54-4, Fig. 1e,g,h,i. Furthermore, overexpression of DDX54 did not restore cell proliferation in cells expressing shDDX54-4, Fig. 1f, indicating that the reduced foci formation observed above could be caused by PKR activation. This finding suggested that shDDX54-4 caused PKR activation through an off-target mechanism, and not through reduced expression of DDX54.
Knockdown of ADAR1 increases PKR activation in combination with the shRNA targeting DDX54
To narrow down the mechanism of PKR activation in cells expressing shDDX54-4, we combined expression of shDDX54-4 with knockdown of ADAR1 in two ADAR1-independent cell lines. These cell lines were chosen because we have previously observed that neither activate dsRNA sensing pathways following knockdown of ADAR1 or DHX9 alone—two proteins shown to suppress activation of dsRNA sensors23,26. We hypothesized that if the effect of shDDX54-4 on PKR activation was dsRNA mediated, that knockdown of ADAR1 could potentially increase PKR activity given ADAR1’s known role in suppression of PKR activation through binding to dsRNA. In support of this hypothesis, we observed that ADAR1 knockdown enhanced PKR activation in cells expressing shDDX54-4, Fig. 2a,b,e,f. Additionally, knockdown of ADAR1 and expression of shDDX54-4 caused reduced foci formation (Fig. 2c,d,g,h). The effects of ADAR1 knockdown on PKR activation in cells expressing shDDX54-4 are consistent with increased activation of PKR due to increased abundance of dsRNA in cells expressing shDDX54-4.
Knockdown of ADAR1 enhances PKR activation in cells expressing shDDX54-4. (a) Representative immunoblot of pPKR and other proteins of interest in SKBR3 expressing shDDX54-4 or shSCR, with or without knockdown of ADAR1. Total protein was imaged using a Stain-Free Gel and was used for normalization. (b) Quantification of pPKR/PKR from the immunoblot in (a). See Supplemental Fig. 2 for quantification of other proteins of interest. (c) Quantification of foci formation shown in (d) for SK-BR-3. (a) Representative immunoblot of pPKR and other proteins of interest in MCF-7 expressing shDDX54-4 or shSCR, with or without knockdown of ADAR1. Total protein was imaged using a Stain-Free Gel and was used for normalization. (b) Quantification of pPKR/PKR from the immunoblot in (a). See Supplemental Fig. 2 for quantification of other proteins of interest. (c) Quantification of foci formation shown in (d) for MCF-7. Bars represent the average of at least three replicates (shown as differently shaped points), error bars are ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. P-values determined by one-way ANOVA with post-hoc Tukey.
shRNA targeting DDX54 activates PKR in vitro
Based on the above data, we hypothesized that shDDX54-4 (and shDDX54-5 which are identical in sequence) may directly activate PKR. To evaluate this, we performed an in vitro kinase assay with purified PKR and shDDX54-4 or shSCR control. As a positive control, we also performed the assay with 106 BLT RNA previously shown to robustly activate PKR27. Unlike shSCR, shDDX54-4 activated PKR in a dose-dependent manner and showed substrate inhibition at higher concentrations as previously observed for PKR, Fig. 3a,b.28These findings are consistent with shDDX54-4 directly activating PKR in cells expressing the shRNA.
(a) Representative PhosphorImage of PKR phosphorylation following in vitro kinase assay with RNAs indicated. (b) Quantification of PhosphorImage shown in (a). Larger points represent the average of at least three replicates (shown as differently shaped smaller points), error bars are ± SD. (c) Predicted structure of shDDX54-4 as determined by mFold41.
Discussion
Activation of dsRNA sensors, such as PKR, has become a promising therapeutic strategy for cancer. For example, depletion of ADAR1, a suppressor of dsRNA sensing, has been shown to promote cell death and anti-tumor immunity in many cellular and in vivo models of cancer23,29,30,31. Many of those studies, and others that focus on different suppressors of dsRNA sensing, have utilized shRNA mediated knockdown to reduce the expression of genes of interest26,32. Here, we report the direct activation of PKR by an shRNA targeting DDX54. We observed activation of PKR by this shRNA in cells constitutively expressing the shRNA and in an in vitro PKR kinase assay. Importantly, our in vitro kinase activity showed substrate inhibition of PKR kinase activity, consistent with previous in vitro studies of PKR activation28. The finding that an shRNA can directly activate PKR brings to light a challenge for studies focused on dsRNA sensing. It is possible, without the correct controls, that experiments that utilize shRNAs may lead to false-positive data indicating that the target gene plays a role in suppression of dsRNA sensing.
Our finding of a shRNA activating PKR is not entirely unique. Early in the use of siRNAs, many groups observed off-target activation of PKR. Off-target activation of PKR has been observed for several different siRNAs targeting multiple genes33,34,35,36,37. Those findings and our own are complicated by biochemical studies of PKR activation that have revealed the requirements for activation of PKR by dsRNA. In vitro studies have shown that a dsRNA of at least 33 bp is required to recruit two PKR monomers and activate PKR, but each dsRBD can independently bind 16 bp length dsRNA20. Recent studies suggest that the minimal length of dsRNA required to activate PKR may change under certain circumstances. Several natural RNA activators of PKR fall below the 33 bp length requirement for PKR activation, such as influenza B ribonucleoprotein (16 bp dsRNA) and interferon-γ mRNA (5–7 bp dsRNA pseudoknot)38,39. Another short RNA element that has been shown to activate PKR is the human immunodeficiency virus 1 (HIV-1) Tat-responsive region RNA (TAR)40. The TAR RNA forms a hairpin structure of similar size to an shRNA, 23 bp, but normally inhibits PKR activation. However, TAR can form intermolecular dimers of sufficient length to activate PKR. Thus, PKR is activated by shorter dsRNAs through multiple mechanisms.
The question remains as to how the shRNA identified here directly activates PKR, while many other shRNAs used by our laboratories and countless other laboratories do not activate PKR. One intriguing aspect of the shRNA is the GC rich sequence near the base of the stem (Fig. 3C). The GC rich region includes seven G-C base pairs, which presumably would stabilize the duplex and thus may contribute to PKR activation. Whether the GC rich stem of the shRNA is important for PKR activation remains to be determined by future experiments.
In the meantime, it is important for researchers to utilize appropriate controls to mitigate the effects of direct activation of PKR (and potentially other dsRNA sensors) by shRNAs. Important measures that can be taken include, (1) Using multiple shRNAs, (2) Performing rescue experiments, (3) Using an orthogonal approach to reduce gene expression, such as knockout by CRISPR-Cas9 or using CRISPR inhibition. It should be noted that when we began these experiments, we did attempt to use multiple shRNAs. We ordered two different shRNA clones from Millipore-Sigma (TRCN0000049957 and TRCN0000289044), both part of the TRC library from the Broad Institute. It was only later that we learned that even though the shRNAs had two different clone identifiers, they were in fact the exact same sequence. We hope that this cautionary tale will help others avoid off-target activation of dsRNA sensors such as PKR and improve the rigor and reproducibility of research in this area.
Materials and methods
Cell culture
Breast cancer cell lines (MCF-7 (RRID: CVCL_0031), SK-BR-3 (RRID: CVCL_0033), BT549 (RRID: CVCL_1092), MDA-MB-231 (RRID: CVCL_0062)) and 293 T (RRID: CVCL_0063) were obtained from American Type Culture Collection. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone) with 10% fetal bovine serum (Bio-Techne), 2 mM glutamine (Hyclone), 0.1 mM nonessential amino acids (Hyclone), 1 mM sodium pyruvate (Hyclone).
Viral production and transduction
Lentivirus was produced by Turbo DNAfection 3000 or LipoFexin (Lamda Biotech) transfection of 293 T cells with pCMV-VSV-G, pCMV-ΔR8.2, and pLKO.1-puro (shSCR and shDDX54) or pLKO.1-hygro (shSCR or shADAR) for shRNAs or pLVX-IRES-Hygro for overexpression of DDX54. Virus was harvested 48 h post-transfection. Cells were transduced with lentivirus for 16 h in the presence of 10 µg/mL protamine sulfate. The cells were selected with puromycin at 2 µg/mL for two days or 100 mg/mL Hygromycin B for five days.
Plasmids
The DDX54 overexpression plasmid (pLVX-IRES-Hygro-FH-DDX54) was generated by PCR amplification of DDX54 from pFRT/TO/FLAG/HA-DDX54, a gift from Markus Landthaler (Addgene plasmid # 97060; http://n2t.net/addgene:97060; RRID: Addgene 97060) and ligation into pLVX-IRES-Hygro. The DDX54 wobble mutant was generated by site-directed mutagenesis using PrimeSTAR Max and In-Fusion cloning (Takara). All primers are available in the Supplemental Information. The lentiviral shRNA plasmids for DDX54 were purchased from Millipore-Sigma, TRCN0000049957 and TRCN0000289044. The shSCR plasmid was a gift from the lab of Sheila Stewart, Washington University in St. Louis.
Immunoblot
Cell pellets were lysed and sonicated in RIPA Buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate and 0.5% sodium deoxycholate) with 1 × HALT Protease Inhibitor (Pierce). Thirty micrograms of protein lysate were resolved on 4–12% TGX Acrylamide Stain-Free gels (Bio-Rad). The Stain-Free gels were imaged prior to transfer to PVDF (Bio-Rad) by TransBlot Turbo (Bio-Rad). The blots were then probed with the appropriate primary antibodies: Primary antibodies: ADAR1 (Santa Cruz, sc-73408; Bethyl Laboratories, A303-883A), DDX54 (Novus Biologicals, NB100-60678), eIF2a (Abcam, ab5369), eIF2a-Ser-51-P (Abcam, ab32157), beta-tubulin (Abcam, catalog no. ab6046, RRID:AB_2210370), cleaved PARP (Cell Signaling Technology, catalog no. 9541, RRID:AB_331426), PKR (Cell Signaling Technology, catalog no. 3072, RRID:AB_2277600), PKR Thr-446-P (Abcam, catalog no. ab32036, RRID:AB_777310). Primary antibodies were detected with horseradish-peroxidase conjugated secondary antibodies (Jackson ImmunoResearch) and detection was carried out with Clarity Western ECL Substrate (Bio-Rad). Densitometry was performed using Image Lab (Bio-Rad). Band intensity was normalized to total protein measured by imaging of the Stain-Free gel.
Foci formation assay
Five thousand cells were plated for each condition in a 10 cm culture dish. After 10 (BT549, MB231 and SK-BR-3) to 20 (MCF-) days the cells were washed briefly with 1 × PBS prior to fixation in 100% methanol for 5 min. After drying, the cells were stained with 0.005% Crystal Violet solution containing 25% methanol (Sigma-Aldrich) prior to washing excess stain away with deionized water. The plates were scanned using an ImageScanner III (General Electric). Foci area was calculated using ImageJ.
Cell proliferation
Three days after transduction with lentivirus for expression of the shRNA listed, 100,000 cells were plated in each well of a 6-well dish. On days two and three post plating, the cells were trypsinized, and resuspended in fresh culture media. Equal volumes of cell suspension and 0.4% Trypan Blue solution were mixed (Cytiva) and the total number of cells in the well was determined using a Bio-Rad TC20 cell counter.
PKR kinase assay
Recombinant PKR was purified from E. coli as described previously27. The PKR kinase assay was performed using purified PKR and either shDDX54-4 (rGr GrCr CrC rGrG rUrG rUrU rCrA rArAr GrGr CrAr UrCr ArUr CrUr CrGr ArGr ArUr GrAr UrGr CrCrU rUrU rGrA rArC rArC rCr GrG rGrU rUr UrU rU) or shSCR (rG rGr UrCr CrUr ArAr GrGrU rUrAr ArGrU rCrGr CrCrC rUrC rGrC rUrCr GrAr GrCrG rArGr GrGr CrGr ArC rUrU rArAr CrCr UrU rAr GrGr UrUr UrU rU), both synthesized by Integrated DNA Technologies as RNA oligonucleotides. The kinase assay was performed as described previously27.
Data availability
Scripts used for data analysis are available here https://github.com/cottrellka/Cottrell-Ryu-et-al-2024. All other data are provided in the main Figures or supplemental Figures and tables.
References
Rehwinkel, J. & Gack, M. U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551. https://doi.org/10.1038/s41577-020-0288-3 (2020).
Hovanessian, A. G. & Justesen, J. The human 2’-5’oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2’-5’ instead of 3’-5’ phosphodiester bond formation. Biochimie 89, 779–788. https://doi.org/10.1016/j.biochi.2007.02.003 (2007).
García, M. A., Meurs, E. F. & Esteban, M. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89, 799–811. https://doi.org/10.1016/j.biochi.2007.03.001 (2007).
Cottrell, K. A., Andrews, R. J. & Bass, B. L. The competitive landscape of the dsRNA world. Mol. Cell 84, 107–119. https://doi.org/10.1016/j.molcel.2023.11.033 (2024).
Reich, D. P. & Bass, B. L. Mapping the dsRNA World. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a035352 (2019).
Shang, R., Lee, S., Senavirathne, G. & Lai, E. C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 24, 816–833. https://doi.org/10.1038/s41576-023-00611-y (2023).
Moore, C. B., Guthrie, E. H., Huang, M. T. & Taxman, D. J. Short hairpin RNA (shRNA): Design, delivery, and assessment of gene knockdown. Methods Mol. Biol. 629, 141–158. https://doi.org/10.1007/978-1-60761-657-3_10 (2010).
Birmingham, A. et al. 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204. https://doi.org/10.1038/nmeth854 (2006).
Jackson, A. L. et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187. https://doi.org/10.1261/rna.25706 (2006).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. https://doi.org/10.1016/j.cell.2004.12.035 (2005).
Meurs, E. F., Galabru, J., Barber, G. N., Katze, M. G. & Hovanessian, A. G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U S A 90, 232–236. https://doi.org/10.1073/pnas.90.1.232 (1993).
Pindel, A. & Sadler, A. The role of protein kinase R in the interferon response. J. Interferon Cytokine Res. 31, 59–70. https://doi.org/10.1089/jir.2010.0099 (2011).
Qiao, H. et al. Cell fate determined by the activation balance between PKR and SPHK1. Cell Death Differ. 28, 401–418. https://doi.org/10.1038/s41418-020-00608-8 (2021).
Ucci, J. W., Kobayashi, Y., Choi, G., Alexandrescu, A. T. & Cole, J. L. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry 46, 55–65. https://doi.org/10.1021/bi061531o (2007).
Su, Q. et al. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 103, 63–68. https://doi.org/10.1073/pnas.0508207103 (2006).
Galabru, J. & Hovanessian, A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J. Biol. Chem. 262, 15538–15544 (1987).
Dey, M., Mann, B. R., Anshu, A. & Mannan, M. A. Activation of protein kinase PKR requires dimerization-induced cis-phosphorylation within the activation loop. J. Biol. Chem. 289, 5747–5757. https://doi.org/10.1074/jbc.M113.527796 (2014).
Nanduri, S., Carpick, B. W., Yang, Y., Williams, B. R. & Qin, J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 17, 5458–5465. https://doi.org/10.1093/emboj/17.18.5458 (1998).
Schmedt, C. et al. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR). J. Mol. Biol. 249, 29–44. https://doi.org/10.1006/jmbi.1995.0278 (1995).
Husain, B., Mukerji, I. & Cole, J. L. Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry 51, 8764–8770. https://doi.org/10.1021/bi301226h (2012).
Kim, Y. et al. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol. Cell 71, 1051-1063.e1056. https://doi.org/10.1016/j.molcel.2018.07.029 (2018).
Cottrell, K. A., Ryu, S., Torres, L. S., Schab, A. M. & Weber, J. D. Induction of viral mimicry upon loss of DHX9 and ADAR1 in breast cancer cells. bioRxiv https://doi.org/10.1101/2023.02.27.530307 (2023).
Kung, C. P. et al. Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer. Oncogene 40, 189–202. https://doi.org/10.1038/s41388-020-01515-5 (2021).
Nicholson, D. W. et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376, 37–43. https://doi.org/10.1038/376037a0 (1995).
Lazebnik, Y. A., Kaufmann, S. H., Desnoyers, S., Poirier, G. G. & Earnshaw, W. C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371, 346–347. https://doi.org/10.1038/371346a0 (1994).
Cottrell, K. A. et al. Induction of viral mimicry upon loss of DHX9 and ADAR1 in breast cancer cells. Cancer Res. Commun. 4, 986–1003. https://doi.org/10.1158/2767-9764.CRC-23-0488 (2024).
Safran, S. A., Eckert, D. M., Leslie, E. A. & Bass, B. L. PKR activation by noncanonical ligands: a 5’-triphosphate requirement versus antisense contamination. RNA 25, 1192–1201. https://doi.org/10.1261/rna.071910.119 (2019).
Kostura, M. & Mathews, M. B. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol. Cell Biol. 9, 1576–1586. https://doi.org/10.1128/mcb.9.4.1576-1586.1989 (1989).
Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102. https://doi.org/10.1038/s41591-018-0302-5 (2019).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48. https://doi.org/10.1038/s41586-018-0768-9 (2019).
Gannon, H. S. et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat. Commun. 9, 5450. https://doi.org/10.1038/s41467-018-07824-4 (2018).
Murayama, T. et al. Targeting DHX9 triggers tumor-intrinsic interferon response and replication stress in small cell lung cancer. Cancer Discov. 14, 468–491. https://doi.org/10.1158/2159-8290.CD-23-0486 (2024).
Zhang, Z., Weinschenk, T., Guo, K. & Schluesener, H. J. siRNA binding proteins of microglial cells: PKR is an unanticipated ligand. J. Cell Biochem. 97, 1217–1229. https://doi.org/10.1002/jcb.20716 (2006).
Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H. & Williams, B. R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5, 834–839. https://doi.org/10.1038/ncb1038 (2003).
Armstrong, M. E. et al. Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J. Immunol. 180, 7125–7133. https://doi.org/10.4049/jimmunol.180.11.7125 (2008).
Puthenveetil, S. et al. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 34, 4900–4911. https://doi.org/10.1093/nar/gkl464 (2006).
Marques, J. T. et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24, 559–565. https://doi.org/10.1038/nbt1205 (2006).
Mayo, C. B., Wong, C. J., Lopez, P. E., Lary, J. W. & Cole, J. L. Activation of PKR by short stem-loop RNAs containing single-stranded arms. RNA 22, 1065–1075. https://doi.org/10.1261/rna.053348.115 (2016).
Kaempfer, R. Interferon-gamma mRNA attenuates its own translation by activating PKR: a molecular basis for the therapeutic effect of interferon-beta in multiple sclerosis. Cell Res. 16, 148–153. https://doi.org/10.1038/sj.cr.7310020 (2006).
Heinicke, L. A. et al. RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 390, 319–338. https://doi.org/10.1016/j.jmb.2009.05.005 (2009).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Acknowledgements
This work was supported by funding to J.D.W from the National Cancer Institute (R01CA262804), funding to B.L.B from the National Institute of General Medical Sciences (R35GM141262), and funding to K.A.C. from the National Institute on Minority Health and Health Disparities (K99MD016946 and R00MD016946).
Author information
Authors and Affiliations
Contributions
K.A.C, S.R., B.L.B. and J.D.W. conceived the project. K.A.C., S.R., J.R.P., H.D., A.A.Y. and H.M. performed the experiments. K.A.C., S.R., J.R.P., H.D., A.A.Y, and H.M. performed the data analysis. K.A.C. and S.R. wrote the manuscript. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cottrell, K.A., Ryu, S., Donelick, H. et al. Activation of PKR by a short-hairpin RNA. Sci Rep 14, 23533 (2024). https://doi.org/10.1038/s41598-024-74477-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-74477-3
Keywords
This article is cited by
-
Leveraging genetics to understand ADAR1-mediated RNA editing in health and disease
Nature Reviews Genetics (2025)