Abstract
Cervical cancer is one of the most commonly diagnosed cancers worldwide, and it is particularly prevalent among women living in developing countries. Traditional classification algorithms often require segmentation and feature extraction techniques to detect cervical cancer. In contrast, convolutional neural networks (CNN) models require large datasets to reduce overfitting and poor generalization. Based on limited datasets, transfer learning was applied directly to pap smear images to perform a classification task. A comprehensive comparison of 16 pre-trained models (VGG16, VGG19, ResNet50, ResNet50V2, ResNet101, ResNet101V2, ResNet152, ResNet152V2, DenseNet121, DenseNet169, DenseNet201, MobileNet, XceptionNet, InceptionV3, and InceptionResNetV2) were carried out for cervical cancer classification by relying on the Herlev dataset and Sipakmed dataset. A comparison of the results revealed that ResNet50 achieved 95% accuracy both for 2-class classification and for 7-class classification using the Herlev dataset. Based on the Sipakmed dataset, VGG16 obtained an accuracy of 99.95% for 2-class and 5-class classification, DenseNet121 achieved an accuracy of 97.65% for 3-class classification. Our findings indicate that DTL models are suitable for automating cervical cancer screening, providing more accurate and efficient results than manual screening.
Similar content being viewed by others
Introduction
Cervical cancer generally affects women and develops from the cervix cells. Cervical is in the lower section of the uterine connected to the vagina. Statistically, it is one of the most commonly occurring cancers affecting women around the world. It is estimated that cervical cancer afflicts over 570,000 women every year and causes more than 311,000 deaths in 20181. In developing countries, cervical cancer cases and deaths account for approximately 85% of all cases and deaths. As of 2030, the World Health Organization (WHO) has established new global targets to lower cervical cancer incidence and death rates by 90%. In addition, it recommends that all countries implement HPV vaccination programs to prevent cervical cancer2.
Additionally, the WHO encourages countries to invest in early detection and treatment of cervical cancer to minimize cervical cancer incidences and mortality. As part of the WHO’s goal to eliminate cervical cancer, each country must provide HPV vaccination to 90% of girls in the age range of 9 to 13 years, as well as provide cervical screening and appropriate treatment to 90% of women in the age range of 30 to 49 years3. During the past decade, high-income countries have implemented organised cervical screening programs that have considerably lowered cervical cancer cases and death rates. The introduction of organised screening programs has enabled early detection and treatment of cervical cancer, resulting in more successful treatment outcomes4,5. The introduction of HPV vaccinations has also contributed to the reduction of cervical cancer cases and rates of death in high-income nations. However, low- and middle-income nations often lack the tools and trained personnel needed to conduct effective cervical screening programs. Additionally, there is often a lack of affordable and accessible HPV vaccinations due to their high cost. As a result, cervical cancer incidence and mortality remain high in these countries.
Cervical cancer screening generally consists of either a Pap smear or a liquid-based cytology test6,7. The Pap smear test includes the collection of cells from the cervix and the examination of these cells under a microscope for possible abnormalities. The liquid-based cytology test involves collecting of cells from the cervix, then analysed under a microscope using a liquid solution. Both tests detect abnormal cells in the cervix that can lead to cancer. Studies have compared the accuracy of Pap tests and liquid-based cytology tests for identifying abnormal cells in the cervix. These studies have found that liquid-based cytology tests are more accurate than Pap tests at detecting abnormal cells. Additionally, liquid-based cytology tests are more comfortable for the patient as the sample collection process is less invasive than a Pap test8. As a result, liquid-based cytology tests are more cost-effective as they require less laboratory processing time and supplies. However, cytology tests have some limitations, including difficulty in interpreting results, which reduce cancer incidence and mortality. Morphological changes in the cytoplasm and nucleus of cells are difficult to detect and require highly skilled personnel to interpret. Furthermore, cytology tests are less sensitive when used early in a cervical cancer diagnosis, as the specimen may not contain abnormal cells6. Additionally, the procedure of collecting a single sample for cytology can be time-consuming and requires experienced personnel. As a result, interpreting results is a labour-intensive process that requires highly trained personnel to identify abnormal cells accurately. Thus, the detection of cervical cancer requires a more accurate and reliable screening method.
To overcome the limitations of manually performed cervical cytology screening, computer vision tools are being used to detect and classify abnormal cells in cervical samples. In computer vision, deep learning algorithms have enabled the more accurate and efficient detection as well as classification of abnormal cells in cervical samples9,10. In addition, these deep learning algorithms can detect and extract features from cervical images with high accuracy, which can reduce the labour-intensive nature of manual screening techniques. Furthermore, deep learning algorithms can identify subtle morphological changes in the cytoplasm and nucleus of cells that may be difficult to detect with manual cytology tests5. In recent years, researchers have developed computer-assisted diagnosis (CAD) systems that categorize single-cell pap smear images or identify irregular cells from full-slide pap smear images. These systems use deep learning algorithms for identification and extraction of features from cervical images with high precision. Manual cytology examinations are often unable to detect subtle morphological changes in the nucleus and cytoplasm of cells. Furthermore, a comprehensive review of computer vision-based cervical cytology screening tools and a comparison of their performance to manual cytology tests can be found in references5,11.
Most studies focus on a single deep-learning model, making it difficult to compare different models. Furthermore, the data sets used in these studies are often small and limited, which may not represent a large population. As such, more research is needed to understand the relative performance of various deep transfer learning models for cervical cancer classification. Different deep learning models have their advantages and disadvantages, which can affect the performance of the cervical cancer classification model. For example, some models may have a better ability to identify certain features in the data, while others may have better generalisation capabilities. In addition, the choice of hyperparameters can also affect the model performance. It is imperative to understand the strengths and weaknesses of various models to select the best model for a given task. In this paper, we examine a performance of sixteen various deep learning models to classify cervical cancer using deep transfer learning. However, such a comprehensive comparison of deep transfer learning models for cervical cancer classification has not been conducted before. Outcome of this study can provide valuable insights for researchers to select the most suitable deep transfer learning model for a given task. Additionally, this study provides a benchmark for future comparisons and can help guide further research into the use of deep transfer learning models for cervical cancer classification.
As a result of this research, the following major contributions are as follows:
-
(1)
To best our knowledge, this is the first research to use 16 different pre-trained models using transfer learning to classify cervical cancer images.
-
(2)
There are two stages of data augmentation used in this research work: the first is used for pre-processing, and the second is applied for in-place data augmentation, using the Keras “ImageDataGenerator” API, where images are reconstructed randomly during training.
-
(3)
For feature extraction, sixteen CNN pre-trained models with enhanced structure, such as VGG16, VGG19, ResNet50, ResNet50V2, ResNet101, ResNet101V2, ResNet152, ResNet152V2, DenseNet121, DenseNet169, DenseNet201, MobileNet, XceptionNet, InceptionV3, and InceptionResNetV2, are introduced.
-
(4)
This study provides significant findings into the performance of various CNN models for classification into multiple classes, in contrast to previous studies that mainly focused on binary classification.
-
(5)
Our proposed techniques achieve the highest classification accuracy score on the Sipakmed dataset in our study.
, The remainder of the paper is organized as follows: Section “Literature work” summarizes relevant conventional CAD methods and deep neural networks for the classification of cervical cancer. Section “Methodology” offers a detailed explanation of database, dataset setting, data augmentation and transfer learning models. Section “Experimental results and analysis” describes the experiment configuration and metrics, and explains the experimental and visual analysis of results. The section “Limitations and future directions” provides insight into the points that need further exploration. Lastly, the section “Conclusion” discusses the outcome of our experiments.
Literature work
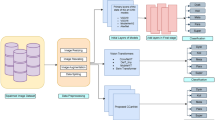
In literature work, three image analysis pipelines have been proposed. These pipelines are shown in Fig. 1. The first two pipelines are traditional techniques that follow different approaches. Traditionally, pipeline-1 relies on hand-crafted features generated from the segmentation of single-cell pap smear images. Using these features, a machine-learning model is trained to detect and analyze single cells within the images. Alternatively, pipeline-2 involves preprocessing of single-cell pap smear images before direct analysis. There are several types of preprocessing, including denoising, image enhancement, and normalization. After pre-processing, the images are fed into a machine-learning model that detects and analyzes single cells. In pipeline-3, which takes advantage of deep convolutional neural networks, involves training a deep learning model to directly analyse the images of single-cell pap smear. Additionally, deep learning has the advantage that they do not require domain experts to design the hand-crafted features for the model to utilise. The model is trained on large datasets of labelled pap smear images and can learn complex features and patterns from the images. Convolutional Neural Networks have been utilized in several previous studies to analyze single-cell pap smear images. Moreover, several deep learning techniques have proven their capacity to automatically diagnose various diseases12,13,14,15 with high accuracy, demonstrating their robustness and reliability. As a result, there is increasing interest in applying deep learning models to improve diagnostic processes and outcomes in medical image analysis.
Related to conventional CAD methods (pipeline 1 or 2)
As mentioned above, conventional CAD methods use a classical ML approach to detect and diagnose abnormalities on pap smear slides. This approach extracts handcrafted features and descriptors from the images. These features are then used to train a model to detect abnormalities. However, this method can be time-consuming. Omneya Attallah16 proposed a CAD model that extracts features from multiple domains by combining three compact deep learning models for high-level spatial features. Additionally, it extracts statistical and textural descriptors from spatial and time-frequency domains for a more accurate representation of cervical cancer characteristics. The model examines the impact of handcrafted attributes on diagnostic accuracy and the effects of merging multiple DL features with handcrafted features. As a result of the quartic SVM, 100% accuracy is achieved. Kalbhor et al.17 the proposed research employed the machine learning based method to detect and distinguish between different types of cervical cells. The DCT and Haar coefficients are extracted, and the feature vectors are fed into seven machine learning algorithms. The results exhibits that the gradient-boosting machine algorithm yields the highest accuracy of 99.47%, followed by random forest, k-nearest neighbour, and naive Bayes with 99.31, 98.74, and 97.12%, respectively. Lavanya Mahmoud et al.18 performed clustering technique segmentation cervical cells and subsequently obtained shape and texture features using the binary histogram Fourier algorithm (BHF). After selecting the features, the quantum grasshopper computing algorithm (QGH) was used as an input to a classifier. Fekri-Ershad et al.19 presented two stage methodology to classify the cervical cancer. Firstly, in the pre-processing step, to minimize noise, enhance features, and normalize them. Then, a multilayer feed-forward neural network is used to detect and analyze single cells in the images. The extracted features from the images were then used as descriptors to be fed to the neural network. The parameters of the neural network were optimised using a genetic algorithm, resulting in an accuracy of 98.9%. Devi et al.20 proposed a modified fuzzy C-means clustering algorithm, extracting geometrical and texture features using Principal Component Analysis (PCA). The KNN classifier achieved an accuracy of 95.8%, while the Fine Gaussian SVM, Ensemble Bagged trees, and Linear Discriminant achieved accuracies of 94.15, 96.28, and 94.86%, respectively. Furthermore, the KNN classifier also showed the least mean squared error of 0.016. This shows that the KNN classification method yields the highest accuracy and lowest error rate for classifying pap smear images into normal and abnormal cells. Chen et al.21 proposed a method that uses two discrete transformation technique to improve the generalisation ability of the TLSE model by combining knowledge learnt from multiple source tasks with snapshot ensemble strategies. Using the fractional coefficient approach, they reduced the dimensionality of the combined features, resulting in a more efficient and accurate classification method. The results witnessed that the reduced features yielded a precision of 81.11% for differentiating between cervical cancer subgroups. Sukumar et al.22 presented a computer-aided automatic method for detecting and diagnosing cervical cancer with the use of Pap smear images. To evaluate the texture and structure of cervical cells, wavelet and GLCM features have been obtained from the cervical images. Based on the extracted features, cervical cell images are classified as normal or abnormal using ANFIS. Based on the results of the proposed method, 92.68% sensitivity, 99.65% specificity and 98.74% accuracy were achieved for dysplastic cell segmentation, demonstrating its accuracy and effectiveness for the detection and diagnosis of cervical cancer. Sarwar et al.23 introduced a hybrid ensemble technique by combining different machine learning algorithms, to improve the accuracy of the proposed method. According to their observation, a hybrid ensemble technique outperforms an individual algorithm, achieving higher accuracy and better classification performance. The results revealed that the hybrid ensemble technique delivered an even higher accuracy of 99.47% in detecting and diagnosing cervical cancer. The system achieved an efficiency of about 96% for the two classes and approximately 78% for the seven classes of cervical cell images.
Related to deep convolutional neural networks (pipeline 3)
Computer-Aided Diagnosis (CAD) systems have been developed to classify, and segment suspicious regions in cervical images. This type of system can provide a more accurate and reliable diagnosis than traditional methods. Furthermore, DL-based CAD systems can facilitate early detection and reduce the time required for diagnosis. Pacal24 presented a framework for evaluating cervical cancer using two publicly available datasets: SIPaKMeD and LBC; both datasets were accurate to 99.02%and 94.48%, respectively. For each model, the author compared the 53 CNN and 53 vision transform models with the SOTA methods. Nitin Kumar Chauhan et al.25 suggested an innovative framework that utilises concatenated features to classify various Pap smear WSI images. The three fine-tuned (VGG-16, ResNet-152, and DenseNet-169) models extract features by performing deep features from the last three layers of each CNN. The extracted features are subsequently concatenated and is used for classification of WSI images. Additionally, the concatenation of features helps to reduce the complexity of the classification process. Suggested model demonstrated a precision of 97.45% for 5-class, and 99.29% for 2-class classifications. This demonstrates the viability of the proposed approach for pap smear WSI images. Attallah26 proposed CAD system “CerCan·Net” for a computerized cervical cancer diagnosis. The model used three lightweight CNNs, with fewer parameters and layers than those commonly used in the literature, which aimed to minimize the complexity of the classification process. CerCan·Net employed transfer learning for the three CNNs, where the pre-trained weights of the models were used to obtain deep features. These weights were then fine-tuned to the task of cervical cancer diagnosis, rather than applying specific deep features to the last three deep layers of each CNN. Additionally, it combines the features acquired using multiple CNN layers. In addition, the study examines the impact of generating a smaller set of deep characteristics that can differentiate different subgroups of cervical cancer using feature selection. The study results showed that CerCan·Net outperformed the ResNet-50 model in terms of accuracy, sensitivity, and specificity. The model achieved an accuracy of 97.7% for the SIPaKMeD dataset and 100% for the Mendeley dataset. Kalbhor et al.27 proposed a hybrid technique combining deep learning architectures with machine learning classifiers for feature extraction from single cell pap smear images and classify them using fuzzy min-max neural networks. Habtemariam et al.28 proposed a deep learning-based approach for automatically detecting the cervix type and classifying cervical cancer. The method incorporated a convolutional neural network (CNN) for the automatic detection of cervix types and a multiple instance learning (MIL) approach for cervical cancer classification. A model developed using MobileNetv2-YOLOv3 was trained and validated to identify the transformation region in the cervix images before extracting the region of interest (ROI) from those images for classification. Using the extracted cervix images, the EffecientNetb0 model was then used to classify the type of cervix using the extracted image. The pre-trained EffecientNetB0 model was then fine-tuned and validated for cervical cancer classification. The results showed that the model could accurately detect the transformation region. Overall, the model provided promising results for the detection and classification of cervical cancer. Park et al.10 compared the machine learning (XGB, SVM, and RF) and deep learning (ResNet-50), to identify signs of cervical cancer. The results showed that the ResNet-50 model yielded an AUC of 0.97, while the average of the three machine learning models yielded an AUC of 0.82. This 0.15-point improvement (p < 0.05) suggests that the ResNet-50 model was also found to have better accuracy, sensitivity, and specificity than the machine learning models, indicating its potential for application in cervical cancer screening. Anurag Tripathi et al.29 used a deep-learning approach to diagnose cervical cancer. They used a CNN-based framework to classify the images from the SIPAKMED dataset into five categories: Dyskeratotic, koilocytotic, metaplastic, parabasal, and superficial intermediate. The CNN model was trained using transfer learning with the VGG-16, ResNet50, and ResNet152 models. The testing precision of ResNet50 was 93.87%, while the ResNet-152 model delivered a precision of 94.89%. VGG-16 achieved the top results with parabasal cells, achieving a precision of 92.85%, while VGG-19 had a slightly higher accuracy of 94.38%. The study by Kudva et al.30 proposed an innovative hybrid transfer learning technique for cervical cancer detection. This technique involves building and training a Convolutional Neural Network (CNN) from scratch, using only the filters identified as relevant by the pre-trained networks AlexNet and VGG-16 Net. The accuracy of the proposed technique was assessed with receiver operating characteristics (ROC) analysis, and an accuracy of 91.46% was achieved using hybrid transfer learning. Ghoneim et al.31 proposed convolutional neural networks (CNNs) based system to classify cervical cancer cells. Image-based CNN models extract deep-learning features from cell images. Afterwards, a classifier based on Extreme Learning Machines (ELM) is used to classify the input images. Herlev database was used to evaluate the performance of the proposed system. The system was trained on 70% of the dataset and tested on the remaining 30%. The system achieved a detection accuracy of 99.5% in the 2-class problem and an accuracy of 91.2% in the 7-class problem.
Table 1 shows a summary of key findings from various studies reviewed in the literature. It highlights the types of datasets used, the methodologies employed, and the high accuracy achieved through various approaches. A significant research gap exists regarding improved methods for detecting and classifying cervical cancer. The review shows that transfer learning leverages pre-trained models to enhance the accuracy and efficiency of cervical cancer detection and classification. Therefore, we conducted a comparative analysis of several pre-trained models to identify the most effective ones. By evaluating their performance metrics, we aim to establish a benchmark that future researchers can use as a reference.
Methodlogy
The aim of this research is to introduce a CAD system using convolutional neural networks for automatic classification of single-cell pap smear images into normal and abnormal cases. Additionally, deep transfer learning can be implemented overcoming the problem of limited data availability because it uses pre-trained models to initialize the weights of a neural network, enabling the model to readily adapt to new data. Fig 2 illustrates the workflow diagram for the proposed method. First, images of cervical Pap smears were gathered from publicly available sources. (Herlev dataset). During the preprocessing step, the implementation of data augmentation is performed to increasing the number of training samples and enhance the model’s generalization capability. Several methods can be employed for this purpose, such as affine transformations, adding noise, canny filters, edge detection, colour filtering, and changing brightness and contrast. Additionally, using the Keras “ImageDataGenerator”, data augmentation can be applied during training time, resulting in a fivefold increase in training samples. Convolutional Neural Network (CNN) is then utilized to extract features from pre-processed image, and to classify it as normal or abnormal. Several state-of-the-art pretrained CNN models were used to evaluate the proposed system, including VGG16, VGG19, ResNet series, InceptionV3, InceptionResNetV2, NASLarge, MobileNet and DenseNet series. Lastly, unseen test images are used to classify normal and abnormal samples and to assess the effectiveness of the proposed system through evaluating its precision, accuracy, recall, and F1.
Schematic representation of the proposed model (Global Average pooling (\(\:{\varvec{G}}_{\varvec{V}\varvec{p}}\)), Batch Normalization \(\:{(\varvec{B}}_{\varvec{N}})\), Dropout layer (\(\:{\varvec{D}\varvec{r}\varvec{o}\varvec{p}}_{\varvec{l}}\)), Dense layer \(\:{(\varvec{D}\varvec{e}\varvec{n}\varvec{s}\varvec{e}}_{\varvec{l}})\), Relu \(\:{(\varvec{R}}_{\varvec{A}\varvec{f}})\) and SoftMax activation functions \(\:{(\varvec{S}}_{\varvec{A}\varvec{f}})\).
Dataset description
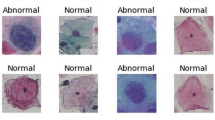
The performance of the proposed framework is evaluated using two publicly available Pap smear datasets. The first dataset is the Herlev dataset, which is available online32, consists of 917 cervical cell images. Table 2 shows the distribution of the Herlev database according to their classes, and Fig. 3 illustrates an example of the collected dataset. The Herlev dataset images are classified into seven classes according to cell morphology and other features33. The Herlev dataset images are classified into seven classes according to cell morphology and other features. These categories include Superficial Squamous (SS), Intermediate Squamous (IS), Columnar Squamous (CS), Mild Dysplasia (MD), Moderate dysplasia (MDD), Severe Dysplasia (SD), and Carcinoma in Situ (CIS) More precisely, Superficial Squamous (SS), Intermediate Squamous (IS), Columnar Squamous (CS) cells are considered normal. Mild Dysplasia (MD), Moderate dysplasia (MDD), Severe Dysplasia (SD), and Carcinoma in Situ (CIS) cell types are abnormal.
The second dataset is the SIPaKMeD dataset, which is available online34. The SIPaKMeD dataset consists of 4049 distinct images of cervical cells. Each image has been cropped from cellular images collected using a charged-coupled device (CCD) camera. The images are further divided into five categories: Parabasal (PB), Superficial-Intermediate (SI), Dyskeratotic (DK), Koilocytotic (KC), and Metaplastic (MP). Additionally, these five groups can be categorized as normal, abnormal, and benign, with the normal group consisting of PB and SI images. An abnormal group contains 1638 images in two categories: DK and KC. MP images are included in the benign category with 793 images. Table 3 shows the distribution of the SIPaKMeD database according to their classes, and Fig. 4 shows some sample of the images of the collected dataset.
Data setting
In this study, 60% of images of each class are used for training, 20% for testing, and the remaining 20% for validation. In addition, data augmentation techniques such as image rotation, flipping, and cropping are applied to the dataset to further enhance the dataset. Thus, the number of images increases by six times for the sipakmed dataset and fourteen times for the herlev dataset, resulting in a more robust and reliable classification. Table 4 presents training, validation, and test datasets for both datasets.
Data augmentation
In this subsection, various data augmentation techniques are performed in our experiment to improve the model’s accuracy. We have used the “imgaug” library, version 4.0, which supports various augmentation techniques such as Affine transformation, Photometric transformation, histogram equalization, Edge detection, Contrast adaptation, and Canny filter. After data augmentation, the training dataset increased by a factor of six. The newly formed images were saved along with the original images.
-
Affine transformation (AT): Affine transformation is a geometric transformation technique that involves scaling, rotating, and translating shearing and horizontal and vertical flip operations of the original images to create updated images. This provides a way to expand the dataset by manipulating the images without losing essential details of the original image.
-
Contrast-limited adaptive histogram equalization (CLAHE): CLAHE is a method that is used to increase the contrast of an image by adjusting the brightness levels of the pixels in the image. All channels CLAHE, CLAHE, and gamma contrast were employed in our experiment to further enhance the contrast of the images. In addition, we increased the variation of the data points in the training dataset. CLAHE was used to adjust the brightness levels of each channel in the image. Gamma contrast was utilized to enhance the image’s contrast by increasing the intensity of the pixels in the image.
-
Edge detection: This module detects edges from random angles and marks the non-edge region as black and the edge region as white. This technique was used to further increase the variation of the data points in the dataset and enable the model to learn more features from the images. We used the directedgedetector imgaug module from the imgaug library to transform the input images into edge images.
-
Canny filter: The Canny filter works by detecting large changes in intensity in the image. It is especially useful for detecting the edges of objects in an image. This allows the model to capture more nuanced features from the images, which can improve the accuracy of the model.
-
Photometric transformations: Photometric transformations can include shuffling all colour channels, turning images into gray-scale and performing colour space transformations. Additionally, it is possible to make changes to the brightness and hue of the image. These techniques can be used to further increase the variation of the data points in the dataset and enable the model to learn more features from the images.
-
Contrast adaptation: This technique increases the contrast of the image, making it easier for the model to learn more features. It can also help the model to better distinguish between objects of similar colours or shades. Additionally, it can help to reduce the effects of noise in the image.
Transfer learning
In this work, transfer learning methods is used to classify the cervical cancer classes. Transfer learning is a deep learning method in computer vision that enables the reuse of knowledge, or transfer, from a pre-trained model to a new task. This method is used to reduce training time and improve the model’s performance by leveraging the knowledge from the pre-trained model. Transfer learning is also advantageous because it requires less data to train the model and can also be used to fine-tune pre-trained models to specific datasets21. The two steps in the transfer learning process are pre-training and fine-tuning. Pre-training involves training a model on a large dataset, such as ImageNet, to obtain a general understanding of the data and features35. For example, a pre-trained deep learning model such as VGG-16 can be used as a pre-trained model for transfer learning. VGG-16 is a convolutional neural network (CNN) that has been trained on the ImageNet dataset and can be used to classify images into 1000 different categories. By leveraging the knowledge from the pre-trained model, VGG-16 can be used to quickly develop a model that can classify images into specific categories with high accuracy. Fine-tuning then involves using the pre-trained model as a starting point and further training it on the specific dataset to adapt it to the new task and improve its performance. For instance, it is necessary to train the entire model from scratch, if we have a large dataset but different from the pre-trained model dataset. In this case, the pre-trained model is not used as the starting point and all the parameters of the model are initialized randomly. This process can be time-consuming and requires a large amount of data, but it can also provide the best performance on the specific task. One of the issues with transfer learning is overfitting36. Overfitting occurs when a model is trained on a dataset that is too small or too like the pre-trained model dataset. In this case, the model may not generalize well and perform as well on the new task. Several techniques can be used to reduce overfitting in transfer learning, such as regularization, data augmentation, and early stopping. Regularization is a technique that constrains the model’s weights to reduce complexity and prevent overfitting. Data augmentation is a technique that is used to increase the size of the dataset by artificially generating new data points. Early stopping is a technique that monitors the model’s performance on the validation dataset and stops the training process when the performance on the validation dataset stops improving. Finally, hyperparameter tuning can be used to adjust the learning rate and other model hyperparameters to achieve the best performance. All these techniques can reduce overfitting and improve the model’s performance on the new task. As a result, the models described below are either used to construct a new algorithm or improve an existing CNN-based model. A summary of selected CNN models with their mathematical expressions can be found in Table 5. Table 5 provides the overview of important parameters and operations of each model. In this table, \(\:{F}_{l}\) denotes as feature maps product at layer \(\:i\), \(\:{y}_{i}\) denotes as input layer \(\:l\), π represent the activation function, \(\:{w}_{i\:}\)denotes the weight and \(\:{b}_{i\:}\)denote as biases.
VGG stands for Visual Geometry Group, developed at the University of Oxford based on CNN technology. It was introduced in 2014 and quickly gained attention for its simple yet effective architecture37. It consists of multiple convolutional layers followed by fully connected layers, with each convolutional layer using small receptive fields (3 × 3 filters) to capture spatial hierarchies. The architecture is known for its simplicity and depth, typically featuring 16 or 19-weight layers38. Due to the large number of parameters in VGG, one of its limitations is that it is costly and consumes a large amount of memory despite its effectiveness. Additionally, the training process can be time-consuming and requires a substantial amount of data and computational power. This study experimented VGG16 and VGG19 with 16 and 19 layers, respectively.
A ResNet is a CNN architecture that contains residual connections between convolutional layers. It was introduced in 2015 by Kaiming He and his colleagues to overcome the problem of vanishing gradients in CNNs38. These residual connections, or skip connections, allow the network to learn residual functions concerning the layer inputs, effectively mitigating the vanishing gradient issue. Additionally, ResNets often utilize bottleneck layers to minimize the model’s dimensions while maximizing its representational power. These bottleneck layers consist of 1 × 1 convolutions that reduce the number of channels, followed by 3 × 3 convolutions, and another set of 1 × 1 convolutions to restore the channel count. This design helps in making the network more efficient without compromising on performance. ResNet is faster and more accurate than its predecessors and is still widely used today. However, ResNet is computationally expensive and requires significant memory resources. As part of this study, we evaluated ResNet50, ResNet50V2, ResNet101, ResNet101V2, ResNet152, and ResNet152V2.
DenseNet is a convolutional neural network architecture developed by Huang et al. in 2017 39. By introducing dense connectivity within layers, DenseNet preserves feature representations and mitigates vanishing gradients, which differs from traditional fully connected layers in CNNs. Rather than connecting all units within each layer, DenseNet only connects adjacent units within each layer. Consequently, DenseNet the number of parameters and increases the receptive field for each unit, resulting in an improved representation of features. There are several variants of DenseNet, including DenseNet-121, DenseNet-169, DenseNet-201, and DenseNet-264, each differing in the number of layers and connections. DenseNet-121 has (6, 12, 24, 16) layers, while DenseNet-169 has (6, 12, 32, 32) layers. DenseNet-201, on the other hand, includes (6, 12, 48, 32) layers, providing different depths and capacities to suit various tasks and datasets. In this study, we evaluate the performance of DenseNet-169, DenseNet-201, and DenseNet-121 model.
MobileNet is designed as a lightweight but powerful network architecture for embedded and mobile systems. It achieves this by using depth-wise separable convolutions, which significantly reduce parameters40. Additionally, MobileNet introduced a shortcut path to bottlenecks, allowing faster computations and improved efficiency. MobileNet variants such as MobileNetV2 and MobileNetV3, which further improve on the original design. In this study, we evaluate MobileNet’s performance.
Inception is introduced in the inception block (also called the inception module), which is a set of convolution layers followed by pooling layers. The Inception block captures features of different sizes and orientations. The use of multiple filters of varying sizes within the same block allows the network to operate at a lower depth while still capturing rich, multi-scale information41. As a result, the model is more efficient and effective at recognizing complex patterns in the data. Inception has several variants, including Inception-v2, Inception-v3, and Inception-v4, each introducing improvements in efficiency and accuracy. In this study, we evaluated the performance of InceptionV3 and InceptionResNetV2.
Xception was introduced by François Chollet at Google42, as an extension of the Inception architecture. Xception relies heavily on Depthwise Separable Convolutions, which reduce the number of parameters and computational costs while maintaining high performance. Another important aspect of optimizing neural networks like Xception is the use of mixed precision training, which helps to speed up training and reduce memory usage. Additionally, mixed precision quantization can be employed to lower inference costs while retaining the model’s accuracy. These techniques are crucial for deploying efficient deep-learning models in resource-constrained environments.
In this paper, we have implemented 16 transfer learning (VGG16, VGG19, ResNet50, ResNet50V2, ResNet101, ResNet101V2, ResNet152, ResNet152V2, DenseNet121, DenseNet169, DenseNet201, MobileNet, XceptionNet, InceptionV3, and InceptionResNetV2) in the transfer learning process, where pre-trained weights are based on ImageNet datasets. ImageNet is a dataset consisting of more than 14 million images in more than 20,000 categories. Firstly, in the transfer learning process, we import all pre-trained models from the Keras library, where we then fine tune the top layer and freeze the convolutional layer to ensure that the weights of the convolutional layers remain unchanged, so that the model parameters are not over-fitted. After the convolutional layer, we add model Global Average pooling (\(\:{G}_{Vp}\)), Dropout layer (\(\:{Drop}_{l}\)), Batch Normalization \(\:{(B}_{N})\), Dense layer \(\:{(Dense}_{l})\) to prevent overfitting. Finally, we feed the output to the SoftMax activation \(\:{(S}_{Af})\)) layer for final classification. We have employed the learning rate of 0.001, the batch size is 32, the number of epochs is 50 and uses ‘’Adam’’ optimizer for minimizing the loss function and training the model. Finally, the proposed models are tested on the test dataset to evaluate its performance.
Experimental results and analysis
Experimental configuration
In this experiment, we have used Google Collaboratory for training and testing the model. It provides a powerful computing resource such as GPUs or TPUs to develop and test models quickly. Furthermore, it is easy to use and free, and comes pre-configured with several machine-learning g libraries including TensorFlow, Matplotlib, Keras, PyTorch, and OpenCV. Moreover, the codes are securely stored in Google Drive.
Performance metrics
To evaluate the performance of the proposed method, we used different performance metrics such as precision, recall, F1 score, and accuracy. Precision metrics \(\:\left({P}_{m}\right)\) measures the ratio of true positives to all positive predictions, while recall metrics \(\:\left({R}_{m}\right)\) measures the ratio of true positives to all actual positive classes. The F1 score \(\:\left({FI}_{s}\right)\) is the harmonic mean of precision and recall, and accuracy metrics \(\:\left({A}_{m}\right)\) measures the ratio of true positives and true negatives to all predictions. Table 3 presents mathematical equations of performance metrics. In Table 3, true positive (\(\:{t}_{p})\) is the number of true positive cases correctly classified, true negative (\(\:{t}_{n})\) is the number of true negative cases correctly classified, false positive (\(\:{f}_{p})\) is the number of false positive cases incorrectly classified, and false negative (\(\:{f}_{n})\) is the number of false negative cases incorrectly classified. In43, a detailed description of the metrics is given and their mathematical formulation is as follows:
Results and analysis
This section presents the results obtained from both CNN-based pre-trained models on two publicly available datasets: SipakMed and HerLev datasets. Various cervical cancer diseases and types are included in this dataset, providing a comprehensive assessment of the model’s performance across different classes of cervical cancer. According to the literature review, data processing involves dividing the dataset into different subsets. Typically, these subsets are used for training, validation, and testing purposes. Therefore, the model not only performs well on unseen data but is also effectively trained. In the current study, the dataset is randomly divided into training, validation, and testing sets with the objective of ensuring the model’s performance is robust and generalizable. Each model required training once, and the most suitable model was selected based on validation data. Following this, this model was tested on unseen testing data. To analyse the performance of fine-tuned models, we evaluated the performance metrics (precision, recall, F1 score, and accuracy) of each model.
Result for the Sipakmed dataset
In this section, we performed a comprehensive comparison between the Convolutional Neural Network (CNN) based fine-tuned models. The main purpose of this work was to evaluate their performance for the classification of cervical cells using the Sipakmed dataset. The comparison results of the fined tuned CNN models for 2-class, 3-class, and 5-class classifications are presented below in detail.
In the 2-class classification, cervical cancer cells are classified into normal (Parabasal (PB), Superficial-intermediate (SI)), and abnormal (Dyskeratotic (DK), Koilocytotic (KC)). The Table 6 illustrates that VGG16 achieved the highest accuracy of 99.95% as well as average precision, recall, and f1-score of 99% among all CNN models. Despite this, MobileNet has the lowest accuracy rate of 96.26%. Models such as VGG19, ResNet50, ResNet50V2, ResNet101, ResNet101V2, ResNet152, ResNet152V2, XceptionNet, InceptionV3, and InceptionResNetV2 achieved intermediate accuracy between 97.61 and 99.95%.
In our 3-class classification, cervical cancer can be classified into three categories: normal, abnormal, and benign, with the normal group consisting of PB and SI images. There are two categories in the abnormal group: DK and KC. The benign group includes MP images. ResNet101V2, ResNet152V, and InceptionV3 demonstrated competitive performance with accuracy values of 93.56, 93.47, and 91.22%, respectively. Both InceptionResNetV2 and NASNetLargeNet achieved intermediate accuracy with 89.10%. In contrast, MobileNet achieved the lowest accuracy of 88.01% among all models. The remaining models, DenseNet121, VGG16, ResNet152, XceptionNet, DenseNet169, VGG19, ResNet50V2, DenseNet201, ResNet50, ResNet101, ResNet101V2, achieved accuracy scores ranging from 93.56 to 97.65%.
In 5-class classification, all classes have been considered. According to Table 6, VGG16 achieved the highest accuracy of 98.66%, followed by VGG19, DenseNet121, ResNet50, DenseNet121, ResNet50, ResNet169, ResNet101, ResNet152, ResNet152V2, InceptionV2, NASNetLargeNet, ResNet101V2, DenseNet201, MobileNet, and ResNet50V2, ranging from 90 to 98% with accuracy of 96.86, 94.57, 94.22, 93.65, 93.56, 92.37, 91.61, 91.34, 91.26, 90.06, 90.01, 90.06, 90.01%, respectively. However, InceptionResNetV2 and XceptionNet achieved the lowest accuracy rates of 89.64 and 65.77%, respectively.
Result for the HerLev dataset
In this section, we compared the pre-trained models based on Convolutional Neural Networks (CNN). This section explains the performance of cervical cell classification using the HerLev dataset. The comparison results of the pre-trained CNN models for 2-class and 7-class classifications are presented below in detail.
In the 2-class classification, cervical cancer cells are classified into normal (Superficial Squamous (SS), Intermediate Squamous (IS), Columnar Squamous (CS)), and abnormal (Mild Dysplasia (MD), Moderate dysplasia (MDD), Severe Dysplasia (SD), and Carcinoma in Situ (CIS)). As shown by Table 7, ResNet50 achieved the highest level of accuracy with 95.10%, followed by ResNet101, VGG16, ResNet101V2, ResNet50V2, VGG19, InceptionV3, DenseNet169, DenseNet121, ResNet152, DenseNet201, ResNet152V2, InceptionResNetV2 XceptionNet, and NASNetLargeNet with an intermediate accuracy score of 93.52, 90.75, 90.51, 90.18, 89.76, 84.73, 84.43, 83.50, 79.44, 73.94, 73.94, 73.29, 71.41, 70.29, and 59.68%, respectively. MobileNet, on the other hand, achieved the lowest accuracy rate of 49.50%.
In 7-class classification problem, all seven classes (Superficial Squamous (SS), Intermediate Squamous (IS), Columnar Squamous (CS), Mild Dysplasia (MD), Moderate Dysplasia (MDD), Severe Dysplasia (SD), and Carcinoma in Situ (CIS)) are considered. Table 7 shows that ResNet50 achieved the best accuracy among all the models tested. The ResNet50V2, InceptionV3, XceptionNet, VGG16, VGG19, ResNet101, ResNet101V2, ResNet152, NASNetLargeNet, and InceptionResNetV2 achieved competitive performance with accuracy ranging from 69.29 to 50.53%. The remaining models achieved accuracy below 50%. It indicates that ResNet50 is the best model among the models tested. Furthermore, the other models can provide useful insights for further optimization.
Visualized analysis
To better understand classification performance, we present comparison of CNN-based pre-trained models on the SipakMed and Herlev dataset in Figs. 5, 6, respectively. The average accuracy of each model is displayed in a dot chart. Nevertheless, it is imperative to consider other factors such as speed, scalability, and memory usage when selecting a CNN-based pre-trained model. Each model has its strengths and weaknesses, and it is imperative to select a model that perfectly fits the application needs.
Moreover, Figs. 7, 8 shows the confusion matrices of some models based on the Sipakmed and Herlev datasets. The Green (light and dark) boxes represent the highest predicted accuracy, while red and orange boxes represent the lowest predicted accuracy.
In Sipakmed dataset, if we examine 2-class classification in Fig. 7a, it can see that VGG16 model achieved 328 correct classification in the normal class and 323 images obtained as abnormal. MobileNet model, on the other hand, achieved the worst performance among all CNN-based pre-trained models, misclassifying 13 images in total. Among the CNN-based models, only the DenseNet121 model achieved 322 correct classifications for the normal class, 322 for the benign class, and 150 for the abnormal class, with only a few misclassifications in Fig. 7b. MobileNet performed not better than its previous performance, achieving 326 correct classifications out of 328 in the normal class and 290 and 111 in the benign and abnormal class, respectively. We can observe in Fig. 7c that the VGG16 model achieved 161 accurate classifications for Parabasal (PB), 167 accurate classifications for Superficial-intermediate (SI), 162 accurate classifications for Metaplastic (MP), and 157 accurate classifications for Dyskeratotic (DK) and Koilocytotic (KC). In contrast, the XceptionNet model achieved the lowest accuracy in five-class classification, with only Superficial-intermediate (SI) achieving 166 correct classifications and one incorrect classification.
In the Herlev dataset, the ResNet50 model performs significantly better in classifying 2-class and 7-class data than the MobileNet model. Figure 8a shows that 130 images were correctly identified as normal and 43 images as abnormal. However, MobileNet was only able to classify 115 images as normal. For 7-class classification, the Resnet50 model achieved 24 correct classifications for Superficial Squamous (SS), 36 correct classifications for Intermediate Squamous (IS), 23 correct classifications for Columnar Squamous (CS), 31 correct classifications for Carcinoma in Situ (CIS), and 14 correct classifications for both Mild Dysplasia (MD), Moderate Dysplasia (MDD) and Severe Dysplasia (SD) respectively. As with the previous dataset, MobileNet did not perform well on the 7-class Herlev dataset as shown in Fig. 8b.
Computational time
In this experiment, we trained and tested individual pre-trained models on the Tesla K80 GPU platform provided by Google Collab. This enabled faster computational times and efficient experiment completion. Table 8 shows that the ResNet50, ResNet50V2, MobileNetV1 and InceptionV3 models had an average computational time of below 55 s per epoch. In contrast, the ResNet152 and ResNet152V2 models had the longest computational time of around 90 s per epoch. Furthermore, the DenseNet series (DenseNet121, DenseNet169, DenseNet201) and VGG series had a computational time of around 60 s per epoch. Finally, the XceptionNet and InceptionResNetV2 models had an average computational time of 65 s per epoch. However, it’s critical to note that these computational times can vary with each execution depending on the specific GPU allocated by Google Collab.
Comparsion with state of the art methods
Cervical cancer disease is detected and classified through several techniques currently available in the literature review; however, in the current study, the performance of the state-of-the-art approaches was compared with that of the developed CNN-based pre-trained method. Table 9 summarizes the performance of the proposed method and the existing state-of-the-art methods in terms of accuracy. The comparison includes details such as the authors and year of publication, the specific method used, the dataset employed, and the accuracy and F1-score achieved by each author.
The authors of Sher Lyn et al.44 developed a transfer learning model with pre-trained CNN models for automated cervical cancer detection with pre-trained CNN models, directly operating on Pap smear images for a seven-class classification task. Thorough evaluation and comparison of 13 pre-trained deep CNN models (VGG-16, VGG-19, DenseNet-121, DenseNet-169, DenseNet-201, ResNet-50, ResNet-101, ResNet-152, Inception, Xception, MobileNet, and MobileNet-v2) were analyzed based on the publicly available Herlev dataset and the Keras package in Google Collaboratory. DenseNet-201 is the best-performing accuracy and performance model. Pacal24 presented a framework for evaluating cervical cancer using two publicly available datasets: SIPaKMeD and LBC; both datasets were accurate to 99.02 ad 94.48%, respectively. For each model, the author compared the 53 CNN and 53 vision transform models with the SOTA methods. In the same period, Nitin Kumar Chauhan et al.45 examined the performance of nine popular deep learning models, namely VGG-16, DenseNet-121, ResNet50, VGG-19, DenseNet-169, Xception, EfficientNetB0, InceptionV3, and ResNet-152 pre-trained on the ImageNet dataset. Using the publicly available Sipakmed dataset, previously trained models are fine-tuned using transfer learning (TL) to classify whole slide pap-smear images (WSI). According to VGG-16, the best DL method can classify 5-classes with 94.89% accuracy and 2-classes with 97.16% accuracy. Several CNN-based models and more than 20 ViT-based models were applied to the SIPaKMeD pap smear dataset in a detailed comparison conducted by Ishak Pacal et al.46. To fine-tune DCNN image classifiers, Mohammed Aliy Mohammed and colleagues47 tested five criteria Pap smears from the SIPaKMeD dataset to test the accuracy of DCNN image classifiers. With an accuracy of 0.990, DenseNet169 exceeded the top ten pre-trained DCNN image classifiers. Anant R Bhatt et al.48 propose a novel method for classifying cervical cells to features extracted from whole slide images. On the Herlev Dataset, the proposed binary and multiclass classification methodology achieved benchmark scores. Comparatively, the proposed approach in 2024 using CNN models achieved a perfect accuracy score of 99.95% on SIPaKMeD dataset, and a 93.52% accuracy value on the Herlev dataset. This method outperforms all previously proposed SOTA methods on these datasets, demonstrating the superiority of the proposed approach in the classification of cervical cancer. In summary, the proposed method of fine tuning pre-trained CNN models demonstrates exceptional performance, achieving perfect accuracy on both the SIPaKMeD and Herlev datasets. As a result, the proposed method demonstrates its efficacy and superiority to existing state-of-the-art methods in the classification of cervical cancer.
Limitations and future directions
In this study, we present a novel algorithm that significantly improves deep-learning applications in cervical cancer classification, it is necessary to acknowledge certain shortcomings. Firstly, the variability in the quality of medical images can affect the algorithm’s performance. Specifically, the analysis of Pap smear images often suffers from overlapping cells and varying staining techniques, making it difficult to identify abnormal cells. These factors can introduce noise and artifacts that make it difficult for the algorithm to identify cancerous cells. Effective preprocessing and enhancement techniques mitigate these issues. In addition, the scarcity of large, annotated datasets poses a challenge for training robust models. This limitation can lead to overfitting, where the algorithm performs well on the training data but poorly on unseen data. In addition, the lack of diversity in the dataset may result in a less generalizable model. Finally, focusing solely on cervical cancer classification may limit the algorithm’s applicability to other classes of cancer and disease. To address this limitation, future work should utilize transfer learning techniques to adapt the model to different classes of cancer and disease. Furthermore, efforts should be made to compile larger, more diverse datasets to improve model robustness and generalizability. Collaborative research with healthcare institutions could also help generate high-quality annotated data, enhancing algorithm performance and reliability.
To improve the accuracy of the CAD technology used for cervical cancer diagnosis, researchers have option to combine CNN’s top models’ classical image features with machine learning knowledge to achieve superior classification performance. Furthermore, the proposed model should be generalized to classify cells that overlap, which reduces the performance of the model. Finally, noise plays a significant role in reducing model performance. To address this problem, techniques such as denoising and restoring images can be implemented. In addition, data enhancement techniques such as image acquisition, rotation and scaling can be applied to further minimize image noise when training deep learning model.
Conclusion
This study proposes sixteen deep learning-based pre-trained CNN models to classify cervical cells using publicly available datasets. This study lays the foundation for further cervical cancer classification research initiatives and evaluates the models in a multi-class cancer classification scenario. The key findings indicate that the proposed models showed high accuracy rates, with some outperforming traditional classification methods. In this study, all pre-trained CNN models are tested on Herlev and Siapkmed datasets, consisting of pap smear single-cell cytopathology images, and achieved the highest accuracy score, except for MobileNet, achieved accuracy levels higher than 60% for both datasets. Using the Herlev dataset, ResNet50 achieved 95% accuracy for both 2-class and 7-class classification tasks. Using the Sipakmed dataset, VGG16 obtained an accuracy of 99.95% for 2-class and 5-class classification, and DenseNet121 achieved an accuracy of 97.65% for 3-class classification. Although our models performed exceptionally well on binary classification tasks with the Sipakmed dataset, the multi-class classification of cancer cells still presents room for improvement. Further optimization and fine-tuning of the models are necessary to enhance their performance in more complex classification scenarios. Future research should aim to address these challenges to achieve even higher accuracy rates across all classification tasks.
Data availability
The following information was provided about data availability: Data are available at the Herlev Cervical Cancer Database (https://mde-lab.aegean.gr/index.php/downloads// ; specifically, Part II: “New Pap-smear Database (images)”).The SIPaKMeD Cervical Cancer Database (https://www.cs.uoi.gr/~marina/sipakmed.html ; data from all cell categories were used).
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Ferlay, J. et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 144, (2018).
Papanicolaou, G. N. New cancer diagnosis. CA Cancer J. Clin. 23, 174–179 (1973).
Elsheikh, T. M. et al. American society of cytopathology workload recommendations for automated pap test screening: developed by the productivity and quality assurance in the era of automated screening task force. Cytopathol 41, 174–178 (2012).
Lozano, R. Comparison of computer-assisted and manual screening of cervical cytology. Gynecol. Oncol. 104, 134–138 (2007).
Davey, E. et al. Effect of study design and quality on unsatisfactory rates, cytology classifications, and accuracy in liquid-based versus conventional cervical cytology: a systematic review. Lancet 367, 122–132 (2006).
Francis, S. A. et al. A qualitative analysis of South African women’s knowledge, attitudes, and beliefs about HPV and cervical cancer prevention, vaccine awareness and acceptance, and maternal-child communication about sexual health. Vaccine 29, 8760–8765 (2011).
Wang, P. et al. Automatic cell nuclei segmentation and classification of cervical pap smear images. Biomed. Signal. Process. Control 48, 93–103 (2019).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Park, Y. R. et al. Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images. Sci. Rep. 11, 16143 (2021).
Hussain, E., Mahanta, L. B., Das, C. R. & Talukdar, R. K. A comprehensive study on the multi-class cervical cancer diagnostic prediction on pap smear images using a fusion-based decision from ensemble deep convolutional neural network. Tissue Cell 65, 101347 (2020).
Pacal, I., Alaftekin, M. & Zengul, F. D. Enhancing skin cancer diagnosis using swin transformer with hybrid shifted window-based multi-head self-attention and SwiGLU-Based MLP. J. Imaging Inf. Med. https://doi.org/10.1007/s10278-024-01140-8 (2024).
Pacal, I., Celik, O., Bayram, B. & Cunha, A. Enhancing EfficientNetv2 with global and efficient channel attention mechanisms for accurate MRI-Based brain tumor classification. Cluster Comput. https://doi.org/10.1007/s10586-024-04532-1 (2024).
Pacal, I. A novel swin transformer approach utilizing residual multi-layer perceptron for diagnosing brain tumors in MRI images. Int. J. Mach. Learn. Cybernet. https://doi.org/10.1007/s13042-024-02110-w (2024).
Kunduracioglu, I. & Pacal, I. Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J. Plant Dis. Prot. 131, 1061–1080 (2024).
Attallah, O. Cervical cancer diagnosis based on multi-domain features using deep learning enhanced by handcrafted descriptors. Appl. Sci. (Switzerland) 13, (2023).
Kalbhor, M., Shinde, S. V. & Jude, H. Cervical cancer diagnosis based on cytology pap smear image classification using fractional coefficient and machine learning classifiers. TELKOMNIKA (Telecommunication Comput. Electron. Control) 20, 1091 (2022).
Mahmoud, H. A. H., AlArfaj, A. A. & Hafez, A. M. A fast hybrid classification algorithm with feature reduction for medical images. Appl. Bionics Biomech. 1–11 (2022).
Fekri-Ershad, S. & Ramakrishnan, S. Cervical cancer diagnosis based on modified uniform local ternary patterns and feed forward multilayer network optimized by genetic algorithm. Comput. Biol. Med. 144, 105392 (2022).
Lavanya Devi, N. & Thirumurugan, P. Cervical cancer classification from pap smear images using modified fuzzy C means, PCA, and KNN. IETE J. Res. 68, 1–8 (2021).
Chen, W., Li, X., Gao, L. & Shen, W. Improving computer-aided cervical cells classification using transfer learning based snapshot ensemble. Appl. Sci. 10, 7292 (2020).
Sukumar, P. & Gnanamurthy, R. K. Computer aided detection of cervical cancer using pap smear images based on adaptive neuro fuzzy inference system classifier. J. Med. Imaging Health Inf. 6, 312–319 (2016).
Sarwar, A., Sharma, V. & Gupta, R. Hybrid ensemble learning technique for screening of cervical cancer using Papanicolaou smear image analysis. Personalized Med. Universe 4, 54–62 (2015).
Pacal, I. & MaxCerVixT: A novel lightweight vision transformer-based approach for precise cervical cancer detection. Knowl. Based Syst. 289, 111482 (2024).
Chauhan, N. K., Singh, K., Kumar, A. & Kolambakar, S. B. HDFCN: A robust hybrid deep network based on feature concatenation for cervical cancer diagnosis on WSI pap smear slides. The Emergence of AI Based Health Informatics 1–17 (2023).
Attallah, O. & CerCan·Net Cervical cancer classification model via multi-layer feature ensembles of lightweight CNNs and transfer learning. Expert Syst. Appl. 229, 120624 (2023).
Kalbhor, M., Shinde, S., Popescu, D. E. & Hemanth, D. J. Hybridization of deep learning pre-trained models with machine learning classifiers and fuzzy min–max neural network for cervical cancer diagnosis. Diagnostics 13, 1363 (2023).
Habtemariam, L. W., Zewde, E. T. & Simegn, G. L. Cervix type and cervical cancer classification system using deep learning techniques. Med. Devices Evid. Res. 15, 163–176 (2022).
Tripathi, A., Arora, A. & Bhan, A. Classification of cervical cancer using deep learning algorithm. In: Proceedings of the Fifth International Conference on Intelligent Computing and Control Systems (ICICCS 2021), Madurai, India Preprint at https://doi.org/10.1109/iciccs51141.2021.9432382 (2021).
Kudva, V., Prasad, K. & Guruvare, S. Hybrid transfer learning for classification of uterine cervix images for cervical cancer screening. J. Digit. Imaging 33, (2019).
Ghoneim, A., Muhammad, G. & Hossain, M. S. Cervical cancer classification using convolutional neural networks and extreme learning machines. Future Gener. Comput. Syst. 102, 643–649 (2020).
Jantzen, J. Pap-smear benchmark data for pattern classification. http://fuzzy.iau.dtu.dk/download/smear2005 (2005).
Herlev Dataset. MDE-Lab. Preprint at https://mde-lab.aegean.gr/index.php/downloads/ (2005).
Plissiti, M. E. et al. Sipakmed: A new dataset for feature and image based classification of normal and pathological cervical cells in pap smear images. 25th IEEE International Conference on Image Processing (ICIP) https://doi.org/10.1109/icip.2018.8451588 (2018).
Maurya, R., Nath Pandey, N., Kishore Dutta, M. & VisionCervix papanicolaou cervical smears classification using novel CNN-vision ensemble approach. Biomed. Signal. Process. Control 79, (2023).
Saini, M. & Susan, S. Cervical cancer screening on multi-class imbalanced cervigram dataset using transfer learning. In: Proceedings – 2022 15th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics, CISP-BMEI 2022 https://doi.org/10.1109/CISP-BMEI56279.2022.9980238 (2022).
Yakkundimath, R., Jadhav, V. S. & Anami, B. S. Automated classification of cervical cells using integrated VGG-16 CNN model. Int. J. Med. Eng. Inf. 16, 185–197 (2024).
Kamal, K. & EZ-ZAHRAOUY, H. A comparison between the VGG16, VGG19 and ResNet50 architecture frameworks for classification of normal and CLAHE processed medical images. https://doi.org/10.21203/rs.3.rs-2863523/v1 (2023).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2261-2269 https://doi.org/10.1109/cvpr.2017.243 (2017).
Xu, T., Li, P. & Wang, X. Cervical lesions classification based on pre-trained mobileNet model. IEEE 15th International Conference on Anti-counterfeiting, Security, and Identification (ASID) https://doi.org/10.1109/asid52932.2021.9651726 (2021).
Dong, N., Zhao, L., Wu, C. H. & Chang, J. F. Inception v3 based cervical cell classification combined with artificially extracted features. Appl. Soft Comput. 93, 106311 (2020).
Chollet, F. Xception: Deep learning with depthwise separable convolutions. 2017 IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR) https://doi.org/10.1109/cvpr.2017.195 (2017).
Grandini, M., Bagli, E. & Visani, G. Metrics for multi-class classification: an overview. (2020).
Tan, S. L., Selvachandran, G., Ding, W., Paramesran, R. & Kotecha, K. Cervical cancer classification from pap smear images using deep convolutional neural network models. Interdiscip. Sci. 16, 16–38 (2024).
Chauhan, N. K., Singh, K. K., Namdeo, S. & Muley, A. An evaluative investigation of deep learning models by utilizing transfer learning and fine-tuning for cervical cancer screening of whole slide pap-smear images. 7th International Conference on Computer Applications in Electrical Engineering-Recent Advances (CERA) 1–5, (2023).
Pacal, I. & Kılıcarslan, S. Deep learning-based approaches for robust classification of cervical cancer. Neural Comput. Appl. 35, 18813–18828 (2023).
Mohammed, M. A., Abdurahman, F. & Ayalew, Y. A. Single-cell conventional pap smear image classification using pre-trained deep neural network architectures. BMC Biomed. Eng. 3, (2021).
Bhatt, A. R., Ganatra, A. & Kotecha, K. Cervical cancer detection in pap smear whole slide images using convNet with transfer learning and progressive resizing. PeerJ Comput. Sci. 7, e348 (2021).
Author information
Authors and Affiliations
Contributions
Harmanpreet Kaur (corresponding author) collected the dataset related to cervical cancer (pap smear images). Dr. Jagroop Kaur assisted with the design of the algorithm and coding. Dr. Reecha Sharma actively participated in the study and performed the statistical analysis. The final version of manuscript has been developed, analyzed, and concluded by Harmanpreet Kaur. Additionally, she assisted in drafting the manuscript. Finally, all authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kaur, H., Sharma, R. & Kaur, J. Comparison of deep transfer learning models for classification of cervical cancer from pap smear images. Sci Rep 15, 3945 (2025). https://doi.org/10.1038/s41598-024-74531-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-74531-0