Abstract
The demand for thermostable pectinase in the global commercial market has expanded progressively at a rapid pace. Many industrial processes are operated at elevated temperatures to break down plant polysaccharides (pectin) owing to the significant use of thermotolerant pectinase. Therefore, the study aimed to optimize the medium constituents for enhanced pectinase production from a thermotolerant pectinolytic bacteria (BIOSMNF45) in combination with the one-variable-at-a-time (OVAT) and response surface methodology (RSM). The isolated BIOSMNF45 with high thermostable pectinase activity was identified as Bacillus siamensis by 16S rRNA sequencing. OVAT technique was used in the preliminary screening of different variables (incubation time, incubation temperature, pH, carbon%, organic nitrogen%, and inorganic nitrogen%) before proceeding to the RSM study. The OVAT result showed that the highest production of thermostable pectinase was attained at 1.00% (w/v) carbon (pectin), 0.40% organic nitrogen (yeast extract), pH 6.00, temperature 45℃ and an agitation of 150 rpm for 24 h incubation. Furthermore, the RSM outcomes revealed that the optimized fermentation conditions boost the thermostable pectinase yield from Bacillus siamensis (BIOSMNF45) by 6.7 folds. It also suggested that the percentage of carbon and temperature showed the highest impact on improved pectinase production compared to other factors.
Similar content being viewed by others
Introduction
The enzyme sector is one of the most essential parts of the manufacturing industry. It was estimated that the global enzyme market would be USD 16.8 billion by 2032, and among this, microbial enzyme accounts for approximately 85.49%1,2. Microbial enzymes are not only a substantial segment of the enzyme market but are the best sustainable and eco-friendly solution to many problems created by using chemicals3. This dominance of microbial enzymes is attributed to the low maintenance and ease of obtaining these enzymes from various microbial sources such as bacteria, fungi, and yeast3. Among all the microbial enzymes produced, the key food enzymes such as amylase, pectinase, lipases, protease, xylanases, etc. have shown much diverse acceptability in various sectors due to their broader action and efficiency4. One such diverse food enzyme is pectinase which breaks down the pectic substance present in the plants. Microbial pectinase has high demand in different commercial sectors because of its ability to break down the pectin present in plant-based raw materials5. The first application of pectinase was made known in the 1930s as a food enzyme in wine manufacturing5. After showcasing its vital role in the food industry, pectinase has taken the limelight and deduced its versatility in multiple domains such as textile, pharmaceutical, cosmetic, and green fuel production5. Microbial pectinases account for more than 10% of the total microbial enzyme market and are expected to grow to USD 26.6 billion by 2030 with 11.10% CARG6. However, the production cost of microbial pectinase is one of the concerns. The raw materials and fermentation media components cover the significant expense in microbial pectinase production7. This factor is crucial in maintaining the quality and quantity of the pectinase produced cost-effectively7. The fundamental aspects of optimization analyses hold immense significance in biotechnology industries, especially when considering the enzyme sector. Research is already being done to optimize the fermentation components to produce high-quality, cost-effective enzymes. Along with the fermentation media, the availability of thermostable pectinase is also of deep interest to researchers for commercial purposes, considering the vast applicability of pectinase8.
The contemporary optimization method of microbial-based product uses a combined approach of response surface methodology (RSM) and one-factor/ variable-at-a-time (OFAT/OVAT). The OFAT has the relevance of being a less complex straightforward, easily understandable, approach9. Despite its simplicity and significance, OFAT cannot be considered the only dependable method for optimization due to its inadequacy in identifying the interaction between the other factors9. Additionally, it is not statistically significant for exploring the entire experimental space broadly10. Therefore, to address these limitations, the most effective way is to introduce a response surface methodology along with OVAT. The most suitable way to combine the principles of science and statistics is to adopt the technique of "design of experiments" (DoE). The introduction of response surface methodology (RSM) in optimization studies completely revolutionized the scientific research and manufacturing sector. The advantage of RSM is that it facilitates the understanding of the relationship between “independent variables” and “dependent variables” by providing a comprehensive and detailed analysis of the system under study11. The advantages of the computational approach are not just limited to interaction study but also give information on robustness, predictability, viability, and precision by fitting a mathematical model of the data for accurate optimization analyses11.
Therefore, considering the significant requirements and demands, this study focuses on optimizing fermentation conditions for enhanced pectinase production from the novel pectinolytic thermotolerant bacteria, Bacillus siamensis (BIOSMNF45), using RSM with D-Optimal quadratic model and is compared with the conventional approach (OVAT).
Materials and methods
Sample collection and isolation of thermotolerant bacteria
Samples from different sources like mud samples of Suraj Kund hot spring, Hazaribagh, and fruit wastes (pulp and peel) were collected from the nearby fruit juice stall of BIT Mesra, Ranchi, Jharkhand, for isolation of various pectin-hydrolyzing bacteria. One gram fruit waste sample and 0.5 g mud sample were dissolved separately and mixed properly in 2 ml autoclaved saline water (0.90% w/v). The mixture was serially diluted till the dilution of 10− 5 and 10− 6, respectively. A volume of 100 µl and 500 µl from each dilution was evenly spread on the sterilized nutrient agar (NA) plates using a sterile L-shape disposable spreader to isolate thermotolerant bacteria. The plates were incubated for 24 h at three different temperatures i.e. 40 ℃, 45 ℃, and 50 ℃. The colonies based on different morphology were picked from the plates and were quadrantally streaked in nutrient agar plates to obtain the pure isolates. The pure isolates were stored in NA plates at 4 ℃ for future experiments.
Screening of pectinolytic isolates
The primary screening of the isolates was done on a pectin screening agar media (PSAM) (pectin 1.00% w/v (NH₄)₂SO₄ 2 g/l, yeast extract 1 g/l, Na₂HPO₄ 6 g/l, KH₂PO₄ 3 g/l, agar 2.50% w/v) by point inoculation method12. The pH of the media before sterilization was adjusted to 5.50 ± 0.50, and the isolates were incubated at 45 ℃ for 48 h. When the growth of the colonies reached about 1–3 mm, the pectinolytic ability of the isolates was checked by Gram’s iodine method. The halo zone around the individual isolates shows the pectin-degrading ability of the pure isolates13. The pectin degrading index (PDI) % and enzymatic index (EI) % were calculated based on the Eqs. (1), (2).
Where HZ represents the diameter of the halo zone and CD represents the diameter of the colony, respectively.
Quantitative estimation of pectinase activity and identification of pectinolytic bacteria
Pectinase activity under standard media ((NH₄)₂SO₄ 0.20%, yeast extract 0.30%, K2HPO4 0.20%, KH2PO4 0.30% KNO3 0.20%, and citrus pectin 1.00%, pH 5.50 at 45 ℃ for 24–72 h in 150 rpm) was measured by the method of Shrestha et al.12 with slight modifications. The liberation of reducing sugars from citrus pectin was quantified by the dinitro salicylic acid (DNSA) method, using D-galacturonic acid as a standard14. The reaction mixture contained 0.1 ml of clarified supernatant as a crude enzyme, 0.2 ml of 1.00% (w/v) citrus pectin dissolved in acetate buffer (0.1 M, pH 6.00), followed by incubation for 15 min at 45 ℃. The reaction was terminated by adding 3 ml of 3,5-dinitro salicylic acid reagent, placed in a 100 ℃ water bath for 10 min, and then cooled in ice water. The release of D-galacturonic acid was measured at 620 nm to quantify pectinase activity. One unit (U/ml) of activity has been defined as the quantity of pectinase needed to release 1 µmol of galacturonic acid per minute (min) at pH 6.00 and 45 ℃. Further, the most potential strain was subjected to Field emission scanning electron microscopy (FESEM) analysis as described by Bibra et al. (2018)15, and sent to the National Center for Cell Science (NCCS) in Pune, India for the 16S rRNA sequenced-based identification.
Optimization of physicochemical parameters using the conventional one-variable-at-a-time approach
The production of pectinase from the novel Bacillus siamensis (BIOSMNF45) under submerged state fermentation (SmF) was optimized by the classical OVAT approach. This approach involves changing one variable/ parameter at a time while maintaining the others at predefined levels/stages. In this work, the selected initial conditions were 24 h incubation, at 45 ℃ at 150 rpm, medium at pH 5.50 with 1.00% w/v carbon source (citrus pectin), 0.20% w/v inorganic nitrogen source ((NH₄)₂SO₄), and 0.30% w/v organic nitrogen source (yeast extract), respectively. The effect of several physicochemical parameters on pectinase synthesis, such as incubation temperature (28− 48 ℃), pH of the production media (5.00–8.00), the concentration of carbon w/v (0.50− 2.00%), the concentration of inorganic nitrogen w/v (0.05− 0.50%), and the concentration of organic nitrogen w/v (0.05− 0.50%), were analyzed. The thermal stability of pectinase was also determined by incubating the enzyme at 45 ℃ for different time intervals (0–120 min). The aliquot of pectinase was withdrawn at specific time intervals, and the enzyme assay was performed. The non-heated enzyme at 0 min was considered a control (100% active). The initial pH values of the production media were adjusted to the desired values before sterilization. The Erlenmeyer’s flasks containing media (50 ml) were autoclaved at a standard condition to maintain sterility. All the experiments were performed in triplicates, and the data were subjected to one-way analyses of variance (ANOVA) carried out using GraphPad Prism software (version 8.0.1.244).
Optimization of physicochemical parameters with response surface model using determinant-optimal quadratic design
The computation approach of RSM using a D-optimal quadratic design was used to optimize the medium for improved pectinase production since it simultaneously estimates the effects of different factors16. The factors and levels used for this optimization (Table 1) were selected by the traditional OVAT method. A polynomial model based on twenty-three experiments with three replicates was built, where the enzyme (pectinase) activity (U/ml) was taken as a response factor (Y) of the designed trials as stated in Table 2 using MODDE 13 (Sartorius Stedim Data Analytics AB, Malmö, Sweden).
The requirement of the number of trials was decided based on Eq. (3):
Where N is the number of experiments, k is the number of factors, and C0 is the central point.
The trial data are subsequently analyzed through the response surface model (RSM) to elucidate the relationship between the independent and dependent variables17. Through Multiple regression analysis (MLR), the data was fitted to a second-order quadratic model according to Eq. (4), which was used to predict the optimal conditions for enhanced pectinase production16.
Where Y represents the response surface, β0 is the intercept (constant), βi, βii, and βij are the linear, quadratic, and interaction coefficients, respectively. At the same time, Xi, Xi2, and XiXj are linear variables, quadratic variables, and interaction of the variables, respectively. The ε is the residual associated (the predicted error) with the experimental trails (represents the difference between measured and predicted R values and evaluates the random variability of our experimental design). The surface plots were generated by altering the values of two factors while maintaining the other factors at their baseline levels. The R2 and the R2predicted values were determined based on Eq. 5 and Eq. 6.
Where R 2 is the determination coefficient, n is the number of measures, i is the number of trials, Ri is the i-th observation value, R’i is the i-th predicted value, and \(\:\overline{R}\) is the average response factor.
Where R 2predicted is the predicted determination coefficient, n is the number of observations, and k is the number of independent variables in the regression equation. The model was simplified by rounding off the non-significant data at a confidence interval of 95%16.
Statistical analysis of the generated model
The experimental trials of the present study were carried out in triplicates, and the values are expressed as mean and standard deviation. All the experiments were performed according to the run order generated by the MODDE software (version 13) to maintain the robustness and unbiasedness of the experiments. The statistical significance was assessed using a one-way analysis of variance (ANOVA), and a p ≤ 0.05 was considered statistically significant for the whole experimental information.
Results and discussion
Isolation and screening of thermotolerant pectinolytic bacteria
During the isolation of bacteria from the mud sample, 06 colonies were grown at 50 ℃. In the case of the fruit waste, 40 bacterial colonies were attained at 45 ℃ and 51 colonies at 40 ℃. Out of which 09 isolates from 40 ℃, 05 isolates from 45 ℃ and 01 isolate from 50 ℃ were selected based on morphological variances for screening pectin utilizing bacteria. The thermotolerant nature of the isolates was ascertained based on Obruča et al. (2022)18, which states that the microbes that required 45 ℃ or above temperature for optimal growth were considered thermotolerant or moderately thermophilic18. Among all the 15 distinct thermotolerant isolates, seven isolates degraded the pectin in the PSAM plate and showed a clear zone of hydrolysis (Table 3). These seven thermotolerant bacteria were considered potent pectinolytic bacteria (primary screening), and were preserved and stored aseptically at 4 ℃. The presence or absence of a particular type of microbes in sampling sites depends upon the physicochemical properties of the surroundings, and the energy source (carbon source) present in that sampling site. Sharma et al.19 isolated the thermophilic pectinolytic bacteria, Bacillus pumilius, from the soil sample at 40 ℃. At the same time, Demir et al.20 isolated Geobacillus steaothermophilus (Ah22) and Brevibacillus borstelenis (P35) at 55–60 ℃ as potential pectinase-producing bacteria from hot spring mud. In the present study, approximately 47% of thermotolerant isolates showed pectinolytic activity when isolated from the waste fruit parts and hot spring mud. Based on the high pectin degrading index (PDI %) and enzymatic index (EI %), three isolates (BIOSMNF45, BIOSMNL02, and BIOSMNM01) were further selected for secondary screening.
Quantitative estimation of pectinase activity and identification of pectinolytic bacteria
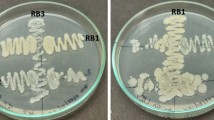
The quantitative estimation of the pectinase activity of selected isolates was carried out by growing them at pH 5.50 at 45 ℃ for 24–72 h. The enzyme activity was estimated every 24 h for quantification of pectinase activity. The isolate BIOSMNF45 yielded the highest pectinase activity as compared to BIOSMNL02 and BIOSMNM01 at 24 h of incubation (Fig. 1). The incubation time is a vital parameter when considering the enzyme activity. In the present study, 24 h of incubation time was found to be the most optimum incubation period for pectinase yield. It shows that either the bacterial culture attains its death phase after 24 h or there is nutrient depletion which could impact bacterial physiology21. Obtaining a high pectinase level in a short incubation period is very interesting, and useful for large-scale production by reducing the possibility of contamination and benefiting the process’s overall economy. The shape and size of the isolate BIOSMNF45 were confirmed by FESEM analysis, which showed that it is a rod shape with a dimension of 1.158 μm length and 561.5 nm width at magnification 100.00 kX (Fig. 2). The 16S rRNA (~ 1200 bp) of the isolate BIOSMNF45 exhibits 99.86% identity with Bacillus siamensis, and hence, the isolate is recognized as Bacillus siamensis (Accession no.: PQ410248). To comprehend the phylogenetic position of strain BIOSMNF45, a phylogenetic tree was constructed based on the isolate’s 16S rRNA sequencing (Fig. 3). Aashish and Bharate have also isolated pectinolytic bacteria from the soil samples and reported the presence of Bacillus sp. as the most dominating pectin hydrolyzing bacterial species22.
Optimization of physicochemical parameters using the conventional one-variable-at-a-time approach
The relevance and cumulative demand for pectinase in different industrial sectors are well-mentioned. In many industrial processes, such as fabric processing, scouring, pulp treatment, bleaching, etc., pectin breakdown was needed in high temperatures13,23. Thermal pectinase is a valuable enzyme in industries where operations often involve elevated temperatures. A specific isolate, i.e., Bacillus siamensis (BIOSMNF45), was selected among all seven isolated pectinolytic strains based on the high thermostable-pectinase production. In this study, we optimized the physicochemical parameters of BIOSMNF45 for enhanced production of thermostable pectinase that can withstand and function efficiently at industrially relevant high temperatures. Under the initial conditions tested, maximal pectinase activity was obtained after one day of incubation at 45 ℃ at pH 5.50. To increase yield, physicochemical conditions such as temperature, pH, carbon (pectin) concentration, inorganic nitrogen ((NH₄)₂SO₄) concentration, and organic nitrogen (yeast extract) concentration were varied. An increase in yield of 85% was observed after the optimization of physicochemical parameters. Figure 4A, B, Figures 5, 6, and Figure 7A, B represent the variations of pectinase activity in the different experiments using the OVAT approach, as explained below.
Effect of incubation temperature on pectinase production
The maximum pectinase activity from strain BIOSMNF45 was found at 45 ℃ (Fig. 4A). According to the literature, temperature has a crucial effect on enzyme production. The production of pectinase was less at lower temperatures, due to the reduced substrate transfer in the cell membrane. In contrast, a temperature higher than the optimum could have denatured the enzyme, resulting in higher energy preservation for cellular growth and lower metabolite generation24. It has also been observed from Fig. 4A that about 80% of pectinase yield is retained at 50 ℃, confirming the thermotolerant nature of B. siamensis (BIOSMNF45). The pre-incubated crude enzyme at 45 ℃ at t = 0 min was considered 100% active (control), and the residual pectinase activity was determined. It was observed from Fig. 4B that pectinase almost retained its original activity for 30 min (99.98% activity retained). There was a drop in stability trend with further incubation because of protein denaturation. However, the enzyme showed stability and retained 78.33% residual activity after 60 min, confirming the thermostable nature of pectinase. Li et al.25 reported higher production of alkaline pectin lyase from B. clausii S-4 at 40 ℃. Similarly, Parmar et al. reported that the optimum temperature for pectinase to hydrolyze the apple pomace is 40 ℃. Whereas Demir et al.,26 have produced alkaline pectin lyase at 50 ℃ in solid-state fermentation from Brevibacillus borstelensis P35. Temperature is an essential physicochemical factor that affects the other parameters of cell growth, and the production of enzymes and metabolites27.
(A) Effect of incubation temperature on pectinase production from B. siamensis (BIOSMNF45), and (B) Thermostability of pectinase from B. siamensis (BIOSMNF45) at 45 ℃. The letters indicate the mean significant difference at p ≤ 0.05, and the error bar represents the standard deviation of triplicate analysis (n = 3), generated using GraphPad Prism software (version 8.0.1.244).
Effect of production media pH on pectinase production
The pH of the production media is a crucial factor that significantly affects the stability of inter-membrane transport, microbial proliferation, and ultimately enzyme production28. Extracellular enzymes are susceptible to pH because they secrete directly into the medium, which is controlled by membrane transport mechanisms that are dependent on the hydrogen ion concentration present in the medium29. To examine the optimum pH for the production of pectinase, the experiments were executed at different pH values ranging from 5.00 to 8.00. Figure 5 shows pectinase production from B. siamensis (BIOSMNF45) which was quite affected by the pH values. In the present work, pectinase activity was obtained in a broad range of pH, showing maximum activity at pH 6.00. Hence, it can be said that the BIOSMNF45 strain produced pectinase in a wide range of pH. While many Bacillus species exhibit peak pectinase in neutral to alkaline environments. In the present study, the optimal pectinase production is at a pH of 6.00-6.80 which is also supported by pectinase production from Bacillus sp., Bacillus sp. MFW7 and Bacillus sphaericus30,31,32.
Effect of carbon (citrus pectin) concentration on pectinase production
The type and percentage of carbon present in the production media for biosynthesis of microbial products is a very critical factor for the optimum growth, metabolism, and yield of products. As pectin acts as an inducer for the production of pectinase, the positive impact was observed by increasing the pectin percentage. The optimum carbon (pectin) concentration was 1.00% (w/v) for the enhanced pectinase production from strain B. siamensis (BIOSMNF45) (Fig. 6). The enzyme production decreases in the higher concentration of citrus pectin, and it might be due to an increase in the viscosity of the fermentation medium, which causes problems and difficulties in maintaining the homogeneity of the fermentation medium and oxygen transfer33. A similar result was reported by Rehman et al.33 for polygalacturonase production from Bacillus licheniformis KIBGE IB-21, where 1.00% pectin as the sole carbon source was the ideal carbon concentration for pectinase activity. Roy et al.34 also reported 1.00% citrus pectin as the most suitable carbon source for boosted pectinase activity among all the other carbon sources used.
Carbon (pectin) % effect on pectinase production from B. siamensis (BIOSMNF45) against the control (absence of pectin). The letters indicate the mean significant difference at p ≤ 0.05, and the error bar represents the standard deviation of triplicate analysis (n = 3), generated using GraphPad Prism software (version 8.0.1.244).
Effect of nitrogen concentration on pectinase production
The concentration of inorganic or organic nitrogen in the fermentation medium has a crucial impact on enzyme synthesis and microbial growth. In the present work, ammonium sulfate was used as an inorganic nitrogen source and yeast as an organic nitrogen source, respectively. The presence of ammonium sulfate had shown an inhibitory effect on the pectinase production from the strain BIOSMNF45 (Fig. 7A) compared to the control. In contrast, the presence of 0.40% (w/v) yeast extract as a sole nitrogen source enhances the pectinase production from strain B. siamensis (BIOSMNF45) (Fig. 7B). A similar report was observed by Govindarajii et al.,35 where 2.00% yeast extract was the most suitable nitrogen source for pectinase production from Streptomyces fumigatiscleroticus VIT-SP4. Prakash et al.36 also reported yeast extract and peptone as better organic nitrogen sources compared to other nitrogen sources studied for pectinase production. Shrestha et al.37 also found organic (peptone) nitrogen source as the better complement for high pectinase activity from Streptomyces sp. Some of the inorganic nitrogen sources such as ammonium sources in the form of sulfate, phosphate, and nitrate showed an inhibitory effect in pectinase production from post-harvested fungi38. Hence, according to previous reports and literature, organic nitrogen sources are more beneficial than inorganic nitrogen sources for pectinase synthesis.
(A) Effect of inorganic nitrogen (ammonium sulphate) % on pectinase production from B. siamensis (BIOSMNF45) against control (absence of ammonium sulphate) (B) Effect of organic nitrogen (yeast extract) % on pectinase production from B. siamensis (BIOSMNF45) against control (absence of yeast extract). The letters indicate the mean significant difference at p ≤ 0.05, the error bar represents the standard deviation of triplicate analysis (n = 3), generated using GraphPad Prism software (version 8.0.1.244).
Optimization of physicochemical parameters with response surface model using determinant-optimal quadratic design
The conventional approach of pectinase production by B. siamensis (BIOSMNF45) indicated that the maximal production was achieved in 24 h of incubation, at pH 6.00, 1.00% (w/v) carbon (citrus pectin), 0.40% (w/v) nitrogen (yeast extract), and at a temperature of 45 ℃ in 150 rpm agitation speed. Taking this condition as a base parameter the independent variables were included in the RSM experiment against the enzyme activity as a response (Y) using a D-optimal quadratic design. The D-optimal quadratic model validated and predicted the condition to produce the maximum pectinase yield. The levels were varied for each variable around the zero level (optimum value determined from the OVAT), the 23 experimental conditions were assayed and the amount of pectinase yielded was recorded in each condition.
The analysis of experimental data generated from the design of the experiment (DoE) using the RSM was assessed by evaluating the raw data, regression analysis, model interpretation, and analyzing the interaction between variables to predict the best parameter point16. The raw data was assessed using condition number, which evaluates the performance of the experimental model before its execution. The condition number should be less than 8 for the “objective of optimization”16; in the present experimental design, it is 6.587. Hence, the performed design is considered a ‘good design’. The model interpretation by regression analysis was done using the summary of the fit plot (Fig. S1) generated by MODDE software (version 13). The R2, Q2, model validity, and reproducibility were evaluated against the response (Y), and the model was fitted with multiple linear regression (MLR)39. The goodness of fit (R2) of 0.996 was obtained in this study, indicating that the model closely fits the raw data. For a good model, the Q2 should be greater than 0.516, and in the present model, the value of Q2 is 0.962 suggesting that the fitted model has outstanding predictability. Reproducibility and model validity give the sense of reliability and suitability of the generated model which should be more than 0.50 and 0.25, respectively16. In the case of the presently studied model, the model validity was 0.62, and the reproducibility was 0.997 which justified the standard values. Thus, it can be said that the demonstrated model is excellently reliable and valid.
The impact or significance of each factor on pectinase production is shown in the coefficient plot (Fig. S2). The factors showing an upbeat in the positive x-axis positively influence the pectinase yield, whereas those showing an upbeat in the negative x-axis indicatea negative influence16. So, it was perceived that the concentration of carbon along with temperature shows the uppermost influence on the yield of pectinase. The second-order polynomial quadratic equation calculated for pectinase production in terms of coded factors is Eq. (7):
Where Y: Pectinase yield (U/ml), X1: carbon percentage, X2: nitrogen percentage, X3: temperature, and X4: pH. The positive sign before the factors indicates a synergetic effect, whereas the negative sign indicates an antagonistic effect on pectinase production. It is preferable to employ the development-based mode with coded components since it can assist in identifying the key variables that can influence the response40. Additionally, to predict the best parameter points, the contour plot (Fig. 8) and the response surface plot (Fig. 9A, B) were generated using MODDE 13 software. The nature of the fitted surface as a maximum, minimum, or “saddle point” is revealed by these plots, which demonstrate the concomitant impact of all four factors on the pectinase yield. The contour plot indicates the interaction and effect of all four factors, where it was found that the yield of pectinase activity increased with the increase in temperature from 42 to 45 ℃ and carbon percentage from 0.50 to 1.00% i.e., a 6.7 folds increase was observed, and it was perceived that the interaction of all four factors has a propitious impact on the yield of pectinase activity . The response surface plot pattern shows the best parameter point of carbon%, nitrogen%, temperature, and pH as variables. The levels around zero (optimum values determined by the OVAT) were found to be the best-fitted points with a synergetic effect as shown in the red zone. Whereas for the levels − 1 & +1, a plodding attenuation in the output was recorded i.e., the yield of pectinase activity was diminutive.
Statistical analysis of the generated model
The analysis of variance (ANOVA) allowed us to determine the relevance of the constructed quadratic model from the experimental dataset (Table 4). ANOVA assessment showed that the constructed D-optimal quadratic model is highly significant with a p-value ≤ 0.000. The F value equals 157.56 implying that the generated model is statistically significant. Within the model, the X1 (carbon%), X3 (temperature), X4 (pH), X12 (carbon%)2, X22 (nitrogen%)2, X32 (temperature)2, X42 (pH)2, X1X2 (carbon% vs. nitrogen%), X2X3 (nitrogen% vs. temperature), X2X4 (nitrogen% vs. pH), and X3X4 (temperature vs. pH) are the significant components with the p-value ≤ 0.05 (Table 4). In contrast, the p-value for the factors X2 (nitrogen%), X1X3 (carbon% vs. temperature), and X1X4 (carbon% vs. pH) are higher than the significant 0.05 value, indicating that these factors and interactions are non-significant. A non-significant lack-of-fit p-value obtained was 0.220, which is higher than the reference value (p ˃ 0.05), and therefore, it can be concluded that the model is not significant relative to the pure error and explained the experimental data correctly41. The adjusted R2 value (0.990) is in agreement with the predicted R2 value (0.996), which validates the proposed model with 99% response variability. Additionally, the low coefficient of variation (CV% = 2.05) indicates that the experimental data is reliable42. A good agreement of “observed versus predicted plot” was generated with low residual standard deviation (RSD) (Fig. S3). In the present study, the low RSD value of 0.02 for the pectinase yield was obtained so, it was assumed that the replicated error is small, and a good model was obtained with a few offset points as indicators of outliners. Overall, it was believed that the outcomes fit the process optimization parameter model reasonably well based on all the analyses.
Conclusion
In the present research, out of all the pectinase-producing bacterial isolates obtained from different sampling sources, BIOSMNF45 (Bacillus siamensis) was the most thermotolerant isolate with maximum thermostable pectinase-producing ability. The OVAT approach showed that maximum pectinase production was achieved at 1.00% (w/v) carbon (pectin), 0.40% nitrogen (yeast extract), and pH 6.00 at 45 ℃ at 150 rpm for 24 h incubation. The presence of an inorganic nitrogen source has shown an inhibitory effect on pectinase production from the strain B. siamensis (BIOSMNF45). The thermostability analysis showed that pectinase maintained its original activity of 99.98% up to 30 min and retained 78.33% residual activity after 60 min of incubation at 45 ℃, confirming the thermostable nature of pectinase. Taking OVAT results as crucial experiments, an RSM approach was used to enhance the thermostable pectinase production and infer the interaction between all the factors. The RSM results showed that thermostable pectinase production increased by 6.7 folds after optimizing production media components. With a R2 value of 0.996, the theoretical result showed strong agreement with the experimental result, indicating that the provided model had an appropriate level of accuracy. Among all the factors (carbon concentration, nitrogen concentration, temperature, and pH) considered, carbon (citrus pectin) concentration and temperature are dynamic in pectinase production. Pectin acts as an inducer of pectinase production and temperature is crucial for the growth and adaptation of B. siamensis (BIOSMNF45) for pectinase synthesis.
Data availability
Availability of data and materials: All data generated or analysed during this study are included in this published article. All information and sources used in the article have been appropriately cited, adhering to copyright guidelines.
References
Expert Market Research. Global Microbial Enzyme Market Report and Forecast 2024–2029. https://www.expertmarketresearch.com/reports/microbial-enzyme-market (2024).
Grand View Research. Microbial Enzyme Market Size, Share & Trends Analysis Report by Type, by Application, by region, and Segment Forecasts, 2024–2030. https://www.grandviewresearch.com/industry-analysis/microbial-enzyme-market (2024).
Singh, R., Kumar, M., Mittal, A. & Mehta, P. K. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 6, 1–15 (2016).
Jan, B., Abass, S. & Ahmad, S. Application of microbial enzymes in Food Industry. In Microbial Biotechnology in the Food Industry: Advances, Challenges, and Potential Solutions. Cham: Springer International Publishing. (323–336). (2024).
Haile, S. & Ayele, A. Pectinase from microorganisms and its industrial applications. Sci. World J. 2022(1), 1881305. (2022).
Zion Market Research. Global microbial enzymes market: Trends, analysis, and forecast. https://www.zionmarketresearch.com/report/microbial-enzymes-market (2024).
Palaniyappan, M., Vijayagopal, V., Viswanathan, R. & Viruthagiri, T. Statistical optimization of substrate, carbon and nitrogen source by response surface methodology for pectinase production using Aspergillus Fumigatus MTCC 870 in submerged fermentation. Afr. J. Biotechnol. 8 (22), 01-09 (2009).
Yu, P., Zhang, Y. & Gu, D. Production optimization of a heat-tolerant alkaline pectinase from Bacillus subtilis ZGL14 and its purification and characterization. Bioengineered 8 (5), 613–623 (2017).
Taylor, C. J., Pomberger, A., Felton, K. C., Grainger, R., Barecka, M., Chamberlain,T. W., … Lapkin, A. A. A brief introduction to chemical reaction optimization. Chem. Rev. 123 (6), 3089–3126. (2023).
Tennalli, G. B. et al. Media optimization for the production of alkaline protease by Bacillus cereus PW3A using response surface methodology. J. Appl. Biol. Biotechnol. 10 (4), 17–26 (2022).
Beg, S., Swain, S., Rahman, M., Hasnain, M. S. & Imam, S. S. Application of design of experiments (DoE) in pharmaceutical product and process optimization. In Pharmaceutical Quality by Design (43–64). Academic. (2019).
Shrestha, S., Khatiwada, J. R., Zhang, X., Chio, C., Kognou, A. L. M., Chen, F., Qin, W. Screening and molecular identification of novel pectinolytic bacteria from forest soil. Fermentation 7 (1), 40. (2021).
Haile, M. & Kang, W. H. Isolation, identification, and characterization of pectinolytic yeasts for starter culture in coffee fermentation. Microorganisms 7 (10), 401 (2019).
Miller, G. L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31 (3), 426–428 (1959).
Bibra, M., Kunreddy, V. R. & Sani, R. K. Thermostable xylanase production by Geobacillus sp. strain DUSELR13 and its application in ethanol production with lignocellulosic biomass. Microorganisms. 6 (3), 93 (2018).
Eriksson, L., Johansson, E., Kettaneh-Wold, N., Wikstrom, C. & Wold, S. Design of Experiments Principles and Applications Umetrics Academy. (2008).
Xie, Y. et al. Collaborative optimization of ground source heat pump-radiant ceiling air conditioning system based on response surface method and NSGA-II. Renew. Energy. 147, 249–264 (2020).
Obruča, S. et al. Polyhydroxyalkanoates synthesis by halophiles and thermophiles: towards sustainable production of microbial bioplastics. Biotechnol. Adv. 58, 107906 (2022).
Sharma, D. C. & Satyanarayana, T. Biotechnological Potential of Agro Residues for Economical Production of Thermoalkali-Stable Pectinase by Bacillus pumilus dcsr1 by Solid‐State Fermentation and Its Efficacy in the Treatment of Ramie Fibres. Enzym. Res. 2012 (1), 281384. (2012).
Demir, N., Nadaroglu, H., Tasgin, E., Adiguzel, A. & Gulluce, M. Purification and characterization of a pectin lyase produced by Geobacillus stearothermophilus Ah22 and its application in fruit juice production. Ann. Microbiol. 61, 939–946 (2011).
Saharan, R. & Sharma, K. P. Production, purification and characterization of pectin lyase from Bacillus subtilis isolated from moong beans leaves (Vigna radiata). Biocatal. Agric. Biotechnol. 21, 101306 (2019).
Aaisha, G. A. & Barate, D. L. Isolation and identification of pectinolytic bacteria from soil samples of Akola region, India. Int. J. Curr. Microbiol. Appl. Sci. 5 (1), 514–521 (2016).
Buchert, J., Pere, J., Puolakka, A. & Nousiainen, P. Scouring of Cotton with Pectinases, Proteases, and Lipases. Text. Chem. Color. Am. Dyest. Report. 32(5) 48 (2000).
Pal, A. & Khanum, F. Production and extraction optimization of xylanase from Aspergillus Niger DFR-5 through solid-state-fermentation. Bioresour. Technol. 101 (19), 7563–7569 (2010).
Li, Z., Bai, Z., Zhang, B., Li, B., Jin, B., Zhang, M., Zhang, H. Purification and characterization of alkaline pectin lyase from a newly isolated Bacillus clausii and its application in elicitation of plant disease resistance. Appl. Biochem. Biotechnol. 167, 2241–2256. (2012).
Demir, N. et al. Purification and characterization of an alkaline pectin lyase produced by a newly isolated Brevibacillus borstelensis (P35) and its applications in fruit juice and oil extraction. Eur. Food Res. Technol. 239, 127–135 (2014).
Mandal, M. & Ghosh, U. Optimization of SSF parameters by OFAT for biosynthesis of cellulase using isolated Aspergillus Niger. Indian J. Chem. Technol. (IJCT). 24 (6), 623–629 (2018).
Xiao, Y., Zhang, X., Zhu, M. & Tan, W. Effect of the culture media optimization, pH and temperature on the biohydrogen production and the hydrogenase activities by Klebsiella pneumoniae ECU-15. Bioresour. Technol. 137, 9–17 (2013).
Bibi, Z., Ansari, A., Zohra, R. R., Aman, A. & Qader, S. A. U. Production of xylan degrading endo-1, 4-β-xylanase from thermophilic Geobacillus stearothermophilus KIBGE-IB29. J. Radiat. Res. Appl. Sci. 7 (4), 478–485 (2014).
Thite, V. S. & Nerurkar, A. S. Physicochemical characterization of pectinase activity from Bacillus spp. and their accessory role in synergism with crude xylanase and commercial cellulase in enzyme cocktail mediated saccharification of agrowaste biomass. J. Appl. Microbiol. 124 (5), 1147–1163 (2018).
Kalaichelvan, P. Production and optimization of pectinase from Bacillus sp. MFW7 using cassava waste. Asian J. Plant. Sci. Res. 2 (3), 369–375 (2012).
Jayani, R. S., Shukla, S. K. & Gupta, R. Screening of bacterial strains for polygalacturonase activity: its production by Bacillus sphaericus (MTCC 7542). Enzym. Res. 2010 (1), 306785. (2010).
Rehman, H. U., Qader, S. A. U. & Aman, A. Polygalacturonase: production of pectin depolymerising enzyme from Bacillus licheniformis KIBGE IB-21. Carbohydr. Polym. 90 (1), 387–391 (2012).
Roy, K., Dey, S., Uddin, M. K., Barua, R. & Hossain, M. T. Extracellular pectinase from a novel bacterium Chryseobacterium indologenes strain SD and its application in fruit juice clarification. Enzym. Res. 2018 (1), 3859752 (2018).
Govindaraji, P. K. & Vuppu, S. Characterisation of pectin and optimization of pectinase enzyme from novel Streptomyces fumigatiscleroticus VIT-SP4 for drug delivery and concrete crack-healing applications: an eco-friendly approach. Saudi J. Biol. Sci. 27 (12), 3529–3540 (2020).
Prakash, H., Chauhan, P. S., General, T. & Sharma, A. K. Development of eco-friendly process for the production of bioethanol from banana peel using inhouse developed cocktail of thermo-alkali-stable depolymerizing enzymes. Bioprocess Biosyst. Eng. 41, 1003–1016 (2018).
Shrestha, S. et al. Optimization of cultural conditions for pectinase production by Streptomyces sp. and characterization of partially purified enzymes. Microb. Physiol. 33 (1), 12–26 (2023).
Rajmane, S. D. & Korekar, S. L. Impact of carbon and nitrogen sources on pectinase production of post-harvest fungi. Current Botany 3(3), 01-03 (2012).
Anuradha, A. & Sampath, M. K. Process optimization for the pretreatment of rice husk with deep eutectic solvent for efficient sugar production. Environ. Technol., 45 (19) ,1–22, (2023).
Montgomery, D. C. Design and Analysis of Experiments Wiley. (2017).
Azzouz, Z. et al. Optimization of β-1, 4-endoxylanase production by an aspergillus Niger strain growing on wheat straw and application in xylooligosaccharides production. Molecules. 26 (9), 2527 (2021).
Pathania, S., Sharma, N. & Handa, S. Optimization of culture conditions using response surface methodology for synergism production of cellulase, xylanase and pectinase by Rhizopus delemar F2 under solid state fermentation. J. Pharmacogn. Phytochem. 6 (6), 1872–1878 (2017).
Acknowledgements
The authors acknowledge UGC-SJSGC under the grant number UGCEC-22-GE-JHA-F-SJSGC-13448. Additionally, the authors express their gratitude for the assistance and resources extended by the Central Instrumentation Facility (CIF), Birla Institute of Technology Mesra in Ranchi, Jharkhand.
Author information
Authors and Affiliations
Contributions
Author Contributions: All authors equally contributed to the conception and design of the present study.The primary conception was planned by [Dr. Vinod Kumar Nigam]. Material preparation, data collection, and analysis were performed by [Anushka Satpathy]. The editing and correction of the manuscript was made by [Dr. Vinod Kumar Nigam], and [Dr. Koel Mukherjee]. Authors consent to the editor for publishing this research article if appropriate.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Satpathy, A., Mukherjee, K. & Nigam, V.K. Improved production of thermostable pectinase from novel Bacillus siamensis (BIOSMNF45) using computational-conventional approach. Sci Rep 15, 11489 (2025). https://doi.org/10.1038/s41598-024-77167-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-77167-2