Abstract
While a broad consensus about the first successful migration modern humans out of Africa seems established, the peopling of Arabia remains somewhat enigmatic. Identifying the ancestral populations that contributed to the gene pool of the current populations inhabiting Arabia and the impact of their contributions remains a challenging task. We investigate the genetic makeup of the current Yemeni population using 46 whole genomes and 169 genotype arrays derived from Yemeni individuals from all geographic regions across Yemen and 351 genotype arrays derived from neighboring populations providing regional context. Principal Component Analysis shows stratification between Yemen districts but also with respect to nearby populations: Yemeni, other Arabian and Bedouin samples form a continuum towards the populations of the Levant, whereas East Africa and India appear strongly differentiated. This finding is further supported by higher Principal Components, admixture and haplogroup analyses, and F-statistics. Moreover, two-reference linkage disequilibrium decay estimates are most significant for Yemeni admixture from an ancient northern influx (up to 5220BP from Palestine) and East Africa (750BP). We show that the initial gene flow into the Yemeni populations of today came from the rest of Arabia and the Levant, and a less substantial and more recent genetic impact into coastal Yemen from East Africa, particularly.
Similar content being viewed by others
Introduction
Yemen has been a leading candidate for one of the Out-Of-Africa expansion routes of modern humans1, with the Sinai Peninsula marking the other2. Subsequently, Yemen remained a major geographical connector for trade and migrations. It has been so far difficult to ascertain when and which of the Out-Of-Africa events that currently dominate the shape of Eurasian settlement may have occurred3. Whether Yemen constituted a permanent habitat for some of these early migrants remains debatable but highly unlikely. During the peak period of the last Ice Age (75-18kya) Yemen and the rest of Arabia were mainly arid, with enough precipitation however to sustain small groups of hunter-gatherers around scattered vegetation in the southern parts of the Arabian Peninsula3,4,5,6,7,8. As the ice started to melt around 18,000 years ago, the Persian Gulf basin started to expand again, flooding the surrounding plains. Coinciding with the basin expanding water levels and the ice melting the entire region from the Levant down to Arabia witnessed a boom of population expansion9. Semi-sedentary hunter-gatherers started to expand from localized habitats along the Caucasus, the Zagros mountains, east Anatolia and the Levant into neighboring populations, along the coasts of the newly shaped Arabo-Persian Gulf and into Arabia7,8. Yemen and the rest of southeast Arabia witnessed additional population influx from the various post-Last Glacial Maximum (LGM) expansions that were occurring throughout the Levant, Anatolia, Mesopotamia and perhaps East Africa.
Subsequently, empires fueled by wealth from wide ranging trade, found Yemen at the crossroads again, connecting the Harappan civilization, Sumer, Dilmun, Egypt, and Mediterranean Levantine cultures such as Israel10, the Phoenicians, Greece and later by British and European mercantilism and colonialism11.
Throughout the last 3 millennia Yemen was a main exporter of incense, myrrh, frankincense and other spices. The trade routes reached as far as the Harappans, and trade from different parts of Africa reaching different parts of Yemen12. Migration between Yemen and East Africa has been very well documented since the second millennium BCE13. In addition to trade of incense, spices and other goods between the old Kingdom of Aksum in Ethiopia, slave trade constituted a major part of moving people from East Africa to Yemen generating population diversity14.
This type of regional diversity can drive the genetic structure differentiating between these regions, offering insight into the broader history15. Previous studies using Y-chromosome analyses have shown that populations within the Arabian Peninsula, including Yemen show some degree of genetic differentiation16,17. Y-chromosome haplogroups and Y-STR markers analyses also demonstrate genetic diversity between Arabia and North Africa on the one hand and between Arabia and the Levant on the other18,19. The Y-haplogroup dispersal, J1 and J1e in particular provides evidence of population movements between the Levant and Arabia dating back multiple millennia19,20. However, mitochondrial DNA analyses present a contrasting perspective showing less mtDNA haplogroup differentiation between Arabian populations and North Africans, but greater difference in mtDNA haplogroup distribution between Arabia and the Levant17,21,22,23. Finally, the combined autosomal DNA, Y-chromosome and mtDNA results provide extensive evidence for dynamic, and multiple back and forth migration events between Arabia and East Africa dating back to at least 12,000 years before present24,25,26,27.
We first sought to examine the impact of back migration from the Levant and Iran on the modern Yemeni gene pool. We also investigated whether the trade of incense, spices and other goods between the old Kingdom of Aksum in Ethiopia and other parts of East Africa resulted in significant human mobility from East Africa to Yemen. Finally, we tested for the presence of the Out Of Africa genetic signatures in modern Yemenis.
Methods
Sample collection and contextualization
Participants were comprehensively recruited from diverse locations in Yemen (Fig. 1). DNA extraction from 3 ml blood was performed in Yemen at Sanaa University. All research was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants.
Map of Yemen. It shows Yemen governorates (black borders). Sampling efforts are shown with scaled triangles at their respective geographic locations. Figure was created using the Python libraries matplotlib and basemap using the basemap method ‘shadedrelie (URL: https://pypi.org/project/basemap/ Version: 1.3.2.).
Genotype array data was obtained for 169 individuals from all governorates of Yemen using the Illumina HumanOmni2.5-8 BeadChip, which comprises 2,338,671 typed loci. Genotyping was conducted at the Wellcome Sanger Institute (UK). Counts, latitude, longitude, sampling locations, including governorate abbreviations are listed in Supplementary Table S1.
We obtained various comprehensive datasets for the purpose of contextualization: the Allen Ancient DNA Resource (AADR/1240 K), 8945 individuals, version 44.328; the Allen Ancient DNA Resource, Human Origins (AADR/HO), 11,686 individuals, version 44.329,30; the Human Genome Diversity Project (HGDP)31 and the East and North Africa (Egypt and Ethiopia, EE)2 and genotype data (Out-Of-Africa, OOA) generated in our lab using the Illumina Human OmniExpress chip32.
The AADR datasets were converted from EIGENSTRAT to binary Plink format. Plink version 1.933 is used to perform the recoding, dataset merging and quality control. We only retained biallelic SNPs and traded off missingness of typed loci for retention of important regional and ancient samples. For F3/F4 statistics and Principal Component Analysis, admixtools were used34. The merged datasets were therefore converted to EIGENSTRAT format using admixtool convertf.
Admixture
For regional contextualization, we excluded samples without latitude and longitude information. For the radii r = (1000, 2000, 3000 km), we retained populations with the majority of their samples within r from the geographic center of Yemen, using the Haversine distance measure (Supplementary Material). We then use Plink to extract only the selected populations for the respective radii. We subjected contextualized datasets to the software tool ADMIXTURE35 using default parameters and running in unsupervised mode for k populations with k = (1,2,…,15) measuring cross validation errors. In addition, we also performed admixture analysis for increasingly larger geographic contexts, with the midpoint in radius r (Supplementary Material).
Principal component analysis (PCA)
We converted datasets to EIGENSTRAT format using convertf. Subsequently we ran smartPCA from EIGENSOFT36. As per general best practice, we excluded ancient samples from the identification of principal components. We used the dataset Human Origins (missingness 0.3, genotype missingness 0.2) for contextualization with ancient samples. We used Kernel Density Estimation from the Python seaborn library in PCA plots, where appropriate.
We also performed a rescaled form of PCA based on bit measurements between individuals’ genomic mutual information using the methods previously described by our group15,37 (Supplementary Material).
F-statistics
We deployed the Admixtools 2.0 library in R4.2 and Admixtools34 for all F-statistics calculations. For measuring population differentiation38 with the Hudson estimator, implemented in is included in Admixtools’ fst function, and we used it for both genotype array and WGS data sets39. To this end we converted the latter from vcf to Plink format using Plink’s recode method. values are visualized in hierarchically clustered heatmaps (R-package heatmap)33. We chose for the merged Human Origins + Yemen genotype dataset the following quality control settings: missingness per individual < 0.3, missingness per genotype < 0.2, Linkage Disequilibrium filtering using Plink and parameters –indep 50 5 2. This approach retains most neighboring populations as well as ancient samples from the region, while maintaining an overlap of 105,232 SNPs, large enough to differentiate admixture in populations from the same continent35 manual Sect. 2.4).
We explored population structures marking admixtures and shared ancestry employing statistics, following Patterson et al.34. The \({F}_{3}\left(C;A,B\right)=\langle \left(C-A\right)\left(C-B\right)\rangle\) will tend to be zero if either \(A\) is uncorrelated with \(B\) and \(C\) is uncorrelated with either \(A\) or \(B\). If \(A\) and \(B\) are correlated and share ancestry not shared by \(C\), then \({F}_{3}\left(C;A,B\right)>0\). To see this, consider an example where \(A=B\). Then \({F}_{3}\left(C;A,B\right)=\langle \left(C-A\right)\left(C-B\right)=\langle {\left(C-A\right)}^{2}\rangle >0.\) Next, if \(C\) contains some admixture from both \(A\) and \(B\), then \({F}_{3}\left(C;A,B\right)<0\). To see this, consider for example, that if \(C=(A+B)/2\) then \({F}_{3}\left(C;A,B\right)=-\langle {\left(B-A\right)}^{2}\rangle /4<0\).
Further, we sought to identify associations within Southwest Asia that may associate with possibly multiple out-of-Africa pathways that passed through the region, and then to identify which associated more strongly with different regions within Yemen. Specifically, for, we constructed two schemas to identify evidence of two pathways out of Africa that may have differentially populated southwest Asia.
First, A was taken as either Sheba or Aden, and B as Lebanese. We selected either Mbuti or chimp as outgroup populations D, against a comprehensive list C (also referred to as X) of all 495 terminal populations (available from HO and our own dataset). Samples with individual missingness < 0.01 and genotype missingness < 0.05 were retained. The top 20 populations with the highest F4 scores, i.e., populations most likely to contribute gene flow into Yemen rather than Lebanon, were presented in the Results section.
We also included Balkans, Turkey, Greece, and Iran and Caucasus (as X) to investigate whether Sheba/Aden more than Lebanon are aligned with western vs. eastern populations. This would provide more information into the negative scale of the F4’s.
We investigated admixture in terms of statistics on both governorate and district level and showed results in forest plots after outlier removal procedure done by smartPCA for districts Sheba and Aden.
In the second schema, B was selected from either Mbuti or Yoruba, and chimp as outgroup, and A was taken as Gujarati (GIH), Iranian, Turkish, and Turkey Arslantepe LateC (late Chalcolithic Anatolia). This group of edges probes lineages to a number of endpoints that likely traveled either through Sinai or Yemen over multiple migrations. Then, D was taken as either Aden or Sheba, and X was selected from several populations drawn from southwest Asia and northeast Africa. To explore differential exposure to populations marking distinct paths out of Africa, populations X were drawn from Southwest Asian and northeast African groups, and D represents Hudaydah, Sheba and Aden.
FineSTRUCTURE
FineSTRUCTURE version 4.1.1 was used to explore various genetic differences among the various Yemeni groups as well as genetic differences between Yemeni and regional populations and we report the results using a fineSTRUCTURE tree40. We used the Yemen Genotype array dataset contextualized with AADR Human Origins (individual missingness 0.3, genotype missingness 0.2), which contains ~ 105 K variants. We reduced the dataset to populations from Central South Asia, Southwest Asia (including Turkey, Caucasus, Levant and Arabia), Sub-Saharan and North Africa, yielding 809 subjects. The dataset was phased using SHAPEIT v5 and the b37 HapMap reference panel.
Whole genome analysis
In addition to Genotyping, Whole Genome Sequencing (WGS) was conducted for 46 samples with > 30 × depth using the Illumina HiSeq X Ten. Genome assembly was done with bwa. The genome analysis pipeline was built with SnakeMake41 (Supplementary Material). The pipeline comprises GRCh38 reference-based genome assembly, base calling, duplicate removal, variant calling and joint genotype calling. It deploys Sentieon version 202,010.0142. Hard variant calling was performed for the sake of comparability when merging with contextual datasets (see below) with bcftools filter command filter -i ’QUAL > 20 && FORMAT/DP > 10’.
We discovered novel SNPs with respect to dbSNP 1.53. The genomes were annotated with ANNOVAR43, which provides allele frequency statistics. For contextualized analyses, we merged the WGS set with the Human Genome Diversity Project44 (HGDP-WGS, 929 samples) and sequences from the Arabian Peninsula45 (APPG, 137 samples) by retaining the intersection of called variants using bcftools: bcftools isec –threads 30 -Oz -p isec -w 1,2,3 -n = 3 and the same filter as above46.
Neanderthal SNP analysis
The merged WGS dataset was subjected to a Neanderthal introgression analysis. Allele frequencies were calculated in all populations for all called variants that were confidently inferred as introgressed from Neanderthal according to the SNP set EUR-ASN from (97,365, SNPs, after intersecting with the set of called variants: 39,057 SNPs)47. Allele frequency vectors are then subjected to hierarchical clustering (UPGMA).
MtDNA Haplogroup identification
Mitochondrial DNA haplogroups were identified by applying haplogrep to the mitochondrial variant calls from the whole genome sequencing analysis48. MtDNA haplogroup frequencies were collected from other studies of nearby populations. Hierarchical clustering (UPGMA) was then applied to these frequency vectors, using the Euclidean Distance as distance measure.
Y-chromosome Haplogroup identification
The final variant calling file from Whole Genome Sequencing was subjected to Y-chromosome haplogroup identification using Y-LineageTracker49.
Admixture linkage disequilibrium decay
ALDER v1.03 was run on the OOA + EE dataset with target population Yemen with setting mindis 0.00550. The weighted linkage disequilibrium was fitted with exponential curve, where n = 24.61 is ALDER’s decay estimate for admixture date, M = 0.00004178 and K = 0.00000378 (and in ALDER output, respectively). Parameter files are provided in the Alder subdirectory of the github repository. We systematically created all possible pairs of reference populations from AMHARA, EGYPT, Egyptian, French, GUMUZ, Iran, OROMO, Palestine, SOMALI, Syria, WOLAYTA and Yoruba. The all-capital designation of some populations in East Africa, are taken from Pagani et al., 20152. The raw output files were parsed and visualized with the Jupyter notebook alderVis.ipynb (see github repository).
Using Yemen as a test population, evidence of multiple admixtures events was inferred with MALDER51. Generation time of 30 was used to derive time of admixtures using non-linkage disequilibrium pruned 215 K SNPs and AMHARA, GUMUZ, SOMALI, OROMO, WOLAYTA, EGYPT, Iran, Iraq, Palestine, Syria, Afghanistan, Lebanon, Jordan, KSA, Turkey and Cyprus as reference populations. A binsize of 0.0005, minimal distance (mindis) of 0.005 Morgan and a maximal distance (maxdis) of 0.5 Morgan was used. Time of admixture does not necessarily indicate target population genetic input into Yemen but time of inferred admixtures of target populations with Yemenis.
Results
Sample collection and contextualization
Sampling efforts are shown in Fig. 1 and Table S1. The results of merging contextual datasets including number of retained samples and SNPs are shown in Table S2. The AADR/1240 K dataset retains highest number of SNPs after merging and therefore yields most accurate F-statistics, while the Human Origin (HO) dataset contains more populations suitable for regional comparison (Table S3).
Admixture
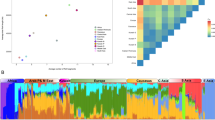
Admixture results with regional contexts within 3000 km are shown for various ancestral populations k = (2…5) in Fig. 2 and k = (2…15) in Supplementary Figure S1 (Fig. S1). East Africa (Ethiopia and Eritrea) differs from Yemen by the substantially stronger Sub-Saharan presence and the absence of an India-specific component (blue for k = 8). Hadramaut and Hudaydah regions differ due to their stronger African component, notably the two governorates with most smartPCA outliers. Their admixture patterns also underlie a greater variability. For each regional context size, the optimal k is determined (Fig. S2).
Admixture (individuals) from populations within 3000 km of Yemen, ordered by geographic location, showing up to K = 5 (Lowest validation error). Hadramaut and Hudaydah regions exhibit strongest diversity. The Yemen governorates were grouped into six districts as follows: Aden (Abyan, Dali, Lahij), Azal (Amran, Dhamar, Saada, Sanaa), Sheba (Bayda, Jawf, Marrib), Hadramaut (Hadramaut, Shabwa), Hudaydah (Hajjah, Hudaydah, Mahwit), Aljanad (Ibb, Taizz).
Principal component analysis
The overall population structure as per Principal Component Analysis, both in global and local contexts, is rather complex, see Fig. 3 (Global PCA), Figure S3 (Yemen District), Figure S4 (Regional PCA within 3000 km) and Figure S5 (Regional PCA within 2000 km).
Global contextualization of Yemen samples A) Yemen forms a cluster (ellipse) and spreads between Levant and Arabian population (towards the bottom). The magnification of this cluster (elliptic inlet, above) shows a complex population structure of Yemen, which is further disentangled by regional contextualization. B) PC1 vs PC3.
The global Principal Component Analysis, performed on the dataset resulting from merging AADR/HO and our samples, is shown in Fig. 3. Yemen samples form a cluster that falls between southwest Asian and North African populations, located on the Sub-Saharan African cline that is commonly observed in global PCA plots.
The rescaled form of PCA shows that PC 1 is dominated by inland Yemeni populations (Sheba, Ibb) with coastal population drawn towards PC 2, see Figure S6.
Regional context
Figure S4 shows clearly accentuated differences between Yemen and the region, with Saudi being closest, while forming a continuum towards the Levant, i.e. reflecting the geography of the Arabian Peninsula (supported by higher PCs). In contrast, there is a substantial gap to East Africa (also visible in the 2000 km context, Figure S5 and the Out-of-Africa context, Figure S7), only sparsely dotted by outliers from coastal Yemen.
Ancient context
Figure S8 shows Yemen samples in the context of ancient samples. The Sheba district that includes samples from Marrib, Jawf, and Bayda, exhibits the greatest similarity to ancient Bronze Age Jordan, Israel and Syria/Ebla.
F-statistics
Yemen governorates bundle closely together in the hierarchical clustering derived from the distance matrix (Fig. S9, Fig. S10). From the compared populations, Egypt, Saudi Arabia and Jewish Yemenite exhibit the highest similarity with Yemen samples. The next-closest cluster comprises wider Southwest Asia, whereas populations from the Indian subcontinent East Africa form remote clusters, thus corroborating PCA results.
F 3 and F 4 statistics
Figure S11 shows \({F}_{3}\left(Aden;Y,GIH\right)\) bar plot, where GIH (Gujarati residing in Houston), and Y are the test cases reported in the bar plot. All the results are positive, except for the Bedouin B group. All the Y values share alleles with GIH that have mutated to the alternate allele in the Yemen population. This suggests that those alleles in Yemen reflect evolution that emerged from an ancient common ancestor to all the other Y’s and the Gujarati. The rest of the tests show a significantly positive \({F}_{3}\) indicating that Gujarati and the \(Y\) share lineages that are not present in Aden.
Figure S12 also shows all positive \({F}_{3}\left(Aden;Y,GIH\right)\), indicating that Gujarati and all the \(Y\)’s share alleles that are different from those in Aden. Figure S13 shows \({F}_{3}\left(Aden;Y,TurkishLateC\right)\), where \(TurkishLateC\) are late Chalcolithic Anatolian aDNA samples, and Y are the test cases reported in the bar plot. All the positive results are southwest Asian, except for the Shaigiya group, who are Sudanese Muslims. The remaining tests show a significantly negative \({F}_{3}\) indicating that Chalcolithic Anatolian and the \(Y\) are admixed into Aden lineages. All of these \(Y\)’s are African. Figure S14 shows \({F}_{3}\left(Aden;Y,Turkish\right)\), where \(Turkish\) are modern Turkish samples, and \(Y\) are the test cases reported in the bar plot. All the results are positive, indicating that all \(Y\) populations share lineages whose mutations are present in modern Turkish lineages and absent in Aden.
All the tested Yemen populations share similar rankings, with coastal exposures showing the strongest African admixtures, but with a long separation between the implied MRCAs, serving to define these populations. The Sheba population is the most isolated, with the most obviously ancient admixture with ancient Anatolia.
The following \({F}_{4}(Sheba/Aden,Lebanon;X,Mbuti/Chimp)\) tests were conducted (First Schema with Mbuti as outgroup, Fig. 4; First Schema with Chimp as outgroup, Figure S15), they show two major groups. The first group with positive F4 scores suggesting more admixture with Sheba than Lebanon. The second group, primarily Central Eurasian, with negative F4 scores favoring more affinity to Lebanon than Sheba.
F4 calculations suggest significant gene flow from Southwest Asian populations, in particular from Bedouins (HGDP group BedouinB, z = 9.87 for outgroup Mbuti) into the Sheba region, while the Aden region is impacted by East Africa (and not significantly by any non-Yemeni Arabian population), though less significantly (Datog and Afar, z-scores between 2.46 and 1.76). Results are robust with respect to outgroups Mbuti and Chimp and are corroborated by the governorate level F4 calculations, shown in Tables S4 and S5, which show that the most significant gene flow happened between landlocked governorates and populations geographically located to the North of Yemen (Bedouin, Saudi). In particular, Marrib shows significant gene flow from Bedouin-B and Saudi Arabia (z-scores 10.635 and 7.39, respectively) (Table S6, Table S7).
The comparison of gene flow between East African (including East African ancient populations) and Lebanese vs. Yemen is shown in Table S8. Remarkably, significant p-scores are only observed for the flow between East Africa and Lebanese. When looking at ancient Eurasian populations, we observe strongly significant impacts on southwest Asian populations only, when compared to Yemen. The latter two include F4 calculations including ancient populations. Likewise, the significant (negative) z-scores in Tables S9 and S10 indicate that southwest Asia populations are more likely to be a recipient of gene flow from the comprehensively tested ancient populations.
The \({F}_{4}(Turkey\_Arslantepe\_LateC,B;X,Aden)\) (Second Schema, Fig. S16) tests for the range of southwest Asian and African populations profiles show, irrespective of A (out-of-Africa destinations, e.g. GIH—Turkey), B (African outgroup), D (Yemen populations), identical rankings of X’s, however the zero-point for F4 shifts. The African populations segregate strongly from the southwest Asian populations but with the Shaigi identifying with Southwest Asians (Fig S16a,b), as expected (recent migrants from the Arabian Peninsula to northern Sudan and surrounding regions).
For F4 (GIH,B;X,Aden), we sought to identify regional differences in the penetration of Indian trade, represented by Gujarati (GIH) and transport into Yemen. In this case, zero represents no correlation with alleles accumulated between X and Aden or RSA and those alleles between Mbuti and GIH. A negative number marks the edge alignment between Aden or RSA and African and southwest Asians (Fig S16c,d).
Haplogroup statistics
Y-chromosomal haplogroups
A phylogenetic tree for Y-chromosomes derived from the 46 fully sequenced genomes is shown in Fig. 5. Haplogroup J1 is predominant throughout Yemen, with high diversity (Fig. S17).
Fig. S18a and b display Y-chromosome haplogroup frequency PCAs for Yemen. The haplogroup frequencies in the wide range PCA (Fig. S18a) show a substantial spread to the Yemeni cluster marking a diversity as large as that of the Arabian Peninsula and Levant combined (Fig. S19).
MtDNA haplogroups
The mtDNA haplogroups derived from the 46 fully sequenced genomes are shown in Table S11-mtDNA Table). Fig. S20 shows the similarity of mtDNA haplogroup frequencies of Yemen compared to nearby populations. The clustering shows that the current study is closest to a previous Yemen study52, Egypt and Saudi Arabia, and, by extension, to North African populations.
The mtDNA Yemeni data (Fig. S18c) are not broken out into Yemen regions. The Yemeni data tend to associate with Egypt and Saudi Arabia.
Linkage disequilibrium decay
Using ALDER, we observe successful tests for admixture events. Amongst the most significant decay fits, the oldest contributions are from Palestine (174 generations, i.e., 5220 ± 1260 BP, z = 8.35), Syria (157 generations), Iran (99 generations) and Egypt (58 generations). Note that the top significant results (Table S12, Figure S21) involve the admixture of ancient populations located to the north of Yemen (Egypt, Iran, Syria, Palestine) and one population from East Africa (Wolayta, Somali).
Using MALDER, we observed consistent results with all previous analyses, showing significant oldest admixture dates of more than 100 generations, with Southwest Asia and Iran (2-reference z-score of 4.02, Table S13, Figure S22) and more recent admixture dates with Ethiopia/Gumuz and Lebanon (2-reference z-score 18.62, Table S14, Figure S23).
FineStructure
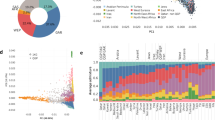
We used iTtol to derive the fineSTRUCTURE tree (Fig. 6)53 and to visually group individuals based on their ancestry. Six main clusters were identified with the majority of Yemeni samples forming one large cluster with few exceptions. The Saudi samples sit closely to the Yemeni cluster while the African samples form a distinct group with one Hadhramauti sample in their midst. Another Hadhramauti group (our data and previously published data) formed a cluster with Moroccan and Mozabite samples.
Whole genome analyses
The variant calling procedure yielded 19,540,858 variants, with the majority being individual mutations (Tables S15, S16, S17, Fig S24). Finally, the analysis of the Neanderthal allele frequencies shows that the Yemenis cluster with samples from the rest of Arabia and the HGDP’s Middle East samples (Fig. S25, Fig. S26).
Discussion
Beyond the broad-brush gradients, establishing Yemen in the context of the Out of Africa Expansion, little remains that is specific within the region, as seen in the ADMIXTURE and PCA plots (Figs. 2, 3, S4, S6 and S7) and supported by F statistics. Yemen sits within the regional gradients of autosomal admixtures.
Yemeni population genetic stratification patterns reveal some regional differences within Yemen. Most Yemeni governorates however exhibit tendencies to form coherent clusters, with relative genetic homogeneity albeit with varying degree of genetic differentiation from other clusters. Among these regions, Hudaydah, Sheba, Aden and Hadramaut show strongly distinguishable stratification patterns, forming tight and distinct clusters in Principal Components Analysis (Fig. S3). The rescaled PCA shows a gradient from interior to coastal Yemen with some African admixture. The difference among Yemeni regions is also reflected in affinity to other regions, as shown in the analyses.
ADMIXTURE plots that resolve at K = 5 show an East African component and a southwest Asian component dominating Yemen with the Hadramaut individuals exhibiting the highest diversity. This is not surprising given that Hadramaut had maritime commerce with India and the entire Arabian Peninsula and had been a British protectorate.
From a regional context, all Yemeni groups lay firmly tied to the base of the African cline in the global PCA plot. Yemen is largely distinguishable from nearby populations, with Saudi Arabian, and HGDP Bedouin groups sampled from the Levantine regions, forming a continuum drawn towards the populations of the Levant. Despite the geographic proximity, we do not observe significant allele sharing between Yemen and East Africa for most of the Yemeni groups tested, with East Africa and India appearing strongly differentiated from Yemen. Surprisingly, Egypt appears to cluster much more closely to Yemen. These findings contrast with conclusions of Richards et al., citing substantial female migration from East Africa into Yemen through slave and female assimilation54,55.
When contextualized with world populations from Sub-Saharan Africa, North Africa, Central South Asia and Southwest Asia, nearly all Yemen samples form a single cluster that also includes the Saudi samples. Consistently, most Yemen samples from Vyas et al., 201752 are subsumed in this cluster. In addition, 8 samples from Hadramaut, that group with 15 samples from Vyas et al., 201752, cluster with Moroccan and Mozabite samples.
Further, Yemen’s Neanderthal allele frequencies show similarities with those from the rest of the Arabian Peninsula and the HGDP-southwestern Asia datasets. These results are also consistent with the narrative placing Yemen within the Arabia Peninsula sphere rather than East Africa with respect to long term admixture and migration. We further note that Yemen samples do not directly overlap with the ancient samples explored.
The Y-haplogroup distribution among the various Yemeni groups is strongly dominated by haplogroup J1 reaching almost 100% in certain regions and the second most frequent Y-haplogroup is E1b. These two haplogroups are the most widely spread Y haplogroups in Arabia and the rest of southwest Asia and are highly suggestive of strong gene flow between these regions and Yemen as observed also in the PCA and ADMIXTURE results. Not only do we observe a high diversity of J1, but it is also present predominantly, in less admixed regions, indicating that it must have dominated the region for a long period. These results add to the body of evidence suggesting that some of these haplogroups have been introduced to Yemen more than 5000 years, hence the level of the observed diversity shown here.
The predominantly African mtDNA L Hg is also well represented in Yemen. Three of the five sequenced samples from Hudayda had a L2a1 mitochondrial haplogroup, the most widespread haplogroup in sub-Saharan Africans. About one third of the Yemeni sequenced samples belonged to the African Hg L suggesting a large African matrilineal influence that paints a different picture from what is observed from the mostly southwest Asian Y Hg predominance in Yemen. These observations are not surprising given the historical trade networks that existed between Africa and Yemen. The African specific mitochondrial DNA haplogroups, most likely reached Yemen from East Africa and remained highly represented in Yemen through isolation. Migration between Yemen and East Africa has been very well documented since the 7th millennium BCE. In addition to trade of incense, spices and other goods between the old Kingdom of Aksum in Ethiopia, slave trade constituted a major part of moving people from East Africa to Yemen.
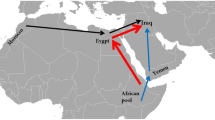
The results from the different Yemen governorates group closely together in the hierarchical clustering with Egypt, Saudi Arabia and Jewish Yemenite exhibiting the highest similarity with Yemen samples. These comparisons are in concordance with our observations for PCA based population stratification. Moreover, two-reference linkage disequilibrium decay analysis shows two significant admixture event estimates. The earliest admixture showing an ancient Levantine influx (Palestine) around 5220BP, noting however that ALDER tends to be more approximate with true dates when recent admixture events are considered. A more recent East African ALDER admixture event around 750BP was observed (Table S11 and Fig S21). MALDER results confirm the older ALDER admixture events providing dates between 100 and 130 generations ago for Levantine-Yemeni Admixtures. It is worth noting however that the older MALDER estimates come with a large variance as expected with increased number of generations. The more recent ALDER admixture event was also supported by the MALDER results. Overall, both MALDER and ALDER analyses indicate earlier Levantine/Southwest Asian admixture with Yemen, and a more recent East African Yemeni admixtures. In essence, these results are consistent with all other observed analyses and support the narrative of an initial gene flow into Yemen from the rest of Arabia and the Levant, followed by more recent genetic input from East Africa and elsewhere.
Impact of more recent admixtures appears in analysis using and statistics, and related methods. \({F}_{3}\) tests show some interesting results, the Shaigiya appear to cluster with SW Asia rather than Africans in Fig S13, supporting their claim to Abbasid descent rather than Nubians. The Chalcolithic Anatolian admixture patterns indicate Aden as a pivotal site between African and Levantine/Arabian Peninsula populations, while the Gujarati tests show penetration that marks Persian/Aryan expansion lineages into modern southwest Asian populations not shared with Aden. The persistence of the general population structure appears strongly in the plots, which rank populations similarly regardless of the details of the reference populations (Fig. S15), where a gap in scores mark the barrier at the Bab el-Mandeb.
Significant gene flow from north Arabian populations, in particular from Bedouins (HGDP group Bedouin-B, z = 9.87 for outgroup Mbuti) into the Sheba region, was strongly suggested using the four-population test (F4) that estimates ancestry proportions (Fig. 4a). The Aden region shows some East Africa ancestry, but to a much lesser impact compared to Sheba and Arabia (Datog and Afar, z-scores 2.46 and 1.61, respectively) results (Fig. 4b). These results were consistent when Chimp was used as an outgroup (Fig S15a and Fig S15b) and were further supported when analyses were performed using governorate level F4 calculations (supplementary Table S4 & Table S5). The Sheba region shows significant gene flow from Bedouin-B and Saudi Arabia (z-scores 10.635 and 7.39, respectively) while the governorate of Bayda, geographically connecting the Aden and Sheba districts received gene flow with almost equal contribution from northern Arabia (particularly Bedouin-B) and East Africa (particularly Afar) with respective z-scores of 4.91 and 4.35. These results are consistent with a continuous gene flow that accompanied post Last Glacial Maximum population expansions from the Levant, through the Arabian Peninsula into the Sheba region.
Vyas et al. previously investigated the population structure in Yemen and demonstrated geneflow between the Levant, Arabia and Yemen52. Our data provide a consistent, yet complementary view on the genetic make-up of the various regions in Yemen and their likely source populations. The Yemenis also show Neanderthal introgression patterns similar to those observed in Bedouins and other southwest Asian populations56.
The limited northern Arabian-Levantine ancestry observed in the southern Yemeni region of Aden was likely diluted by the flow from East Africa. The findings from Bayda clearly illustrates the observed gradient: a northern Arabian-Levantine meeting a southern East African genetic gradient. For decades, the prevailing literature57 has attributed cultural and linguistic similarities between the Levant and Yemen to multiple south-north population migration waves, from Yemen to the coastal Levant, particularly in the aftermath of the bursting of the Marrib dam during the second of third Century CE. But recent evidence by Al Jallad and others document an (eastern) Levantine genesis of “classical” Arabic supporting the narrative presented here and previous arguments proposing cultural and linguistic flow from the Levant to Yemen. These findings are also supported through the Bayesian phylogenetic analysis by Kitchen et al. estimating an Early Bronze Age origin for Semitic languages in the Levant58,59,60.
Our results paint a picture of Yemen being in a crossroads, marked by the large-scale baseline genetic diversity that largely evolved many millennia. Yemen witnessed an initial gene flow pattern defined by the more ancient markers from the rest of the Arabian Peninsula and the Levant that started as early as the Epipaleolithic period followed by population isolation and limited expansion. Yemen witnessed additional, but less substantial, flow from southwest Asia, Jordan and Syria through the Nabateans during the active incense trade between the Levant and Yemen, from Arabia during the Islamic Expansion, and a subsequent one from the west and southwest dominated by East Africa. The other subsequent input phase came though East Africa and may have started around the 7th Millennium BCE with the onset of the warmer and wetter conditions of the Holocene. A period that has witnessed major population expansions throughout the region. Additional and substantial gene flow from East Africa into Yemen likely reflects persistent trade from ancient Egypt to modern times, as reflected by numerous archaeological remains, and historical periods, such as the second and sixth century CE Kingdom of Aksum that was established in northern Ethiopia, then invaded and settled in Yemen.
Crossroads tended to do well, until, like Yemen, they were bypassed. Yemen retains a genetic legacy that reveals the scope of her connections in remarkable detail over the years where there was no choice but to deal with her for transportation of these precious commodities.
Data availability
Whole Genome Sequencing data of 46 samples is available at the European Genotype–Phenotype Archive (EGA) under accession number EGAD00001003097. Code Availability All code referred to in the manuscript as well as interactive html/javascript pages of PCA plots are available at https://github.com/HenschelLab/YemenPopGen.
Code availability
All code referred to in the manuscript as well as interactive html/javascript pages of PCA plots are available at https://github.com/HenschelLab/YemenPopGen.
References
Macaulay, V. et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308 (5724), 1034–1036 (2005).
Pagani, L. et al. Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am. J. Hum. Genet. 96 (6), 986–991 (2015).
Beyer, R. M., Krapp, M., Eriksson, A. & Manica, A. Climatic windows for human migration out of Africa in the past 300,000 years. Nat. Commun. 12 (1), 4889 (2021).
Groucutt, H. S. et al. Human occupation of the Arabian Empty Quarter during MIS 5: evidence from Mundafan Al-Buhayrah, Saudi Arabia. Quat. Sci. Rev. 119, 116–135 (2015).
Breeze, P. S. et al. Palaeohydrological corridors for hominin dispersals in the Middle East ∼250–70,000 years ago. Quat. Sci. Rev. 144, 155–185 (2016).
Groucutt, H. S., Scerri, E. M. L., Stringer, C. & Petraglia, M. D. Skhul lithic technology and the dispersal of Homo sapiens into Southwest Asia. Quat. Int. 515, 30–52 (2019).
Petraglia, M. D., Breeze, P. S. & Groucutt, H. S. Blue Arabia, Green Arabia: Examining human colonisation and dispersal models. In Geological Setting, Palaeoenvironment and Archaeology of the Red Sea (eds Rasul, N. M. A. & Stewart, I. C. F.) 675–683 (Springer International Publishing, 2019).
Rose, J. New light on human prehistory in the Arabo-Persian Gulf oasis reply. Curr. Anthropol. 51, 849–883 (2010).
Heller, J. Near East Ecosystems, Animal Diversity (Elsevier, 2001).
Laursen, S. T. et al. From Dilmun to Wādī al-Fāw: A forgotten desert corridor, c. 2000 BC. Arab. Archaeol. Epigr. 34 (1), 63–86 (2023).
During Caspers ECL, Olijdam E, Spoor RH. Intercultural relations between south and southwest Asia : studies in commemoration of E. C. L. During Caspers (1934–1996): Oxford: Archaeopress; (2008).
Copeland P, Bird J, Blue L, Gupta S, James S, Sedov A, et al. Food for the Gods New Light on the Ancient Incense Trade. Peacock D, Williams D, editors: Oxbow Books; (2007).
Phillipson, D. W. Relations between southern Arabia and the northern Horn of Africa during the last millennium BC. Proc. Semin. Arab. Stud. 41, 257–265 (2011).
Gilboa, A. & Namdar, D. On the beginnings of South Asian spice trade with the Mediterranean region: A review. Radiocarbon 57 (2), 265–283 (2015).
Platt, D. E. et al. Autosomal genetics and Y-chromosome haplogroup L1b–M317 reveal Mount Lebanon Maronites as a persistently non-emigrating population. Eur. J. Hum. Genet. 29 (4), 581–592 (2021).
Alshamali, F., Pereira, L., Budowle, B., Poloni, E. S. & Currat, M. Local population structure in Arabian Peninsula revealed by Y-STR diversity. Hum. Heredity 68 (1), 45–54 (2009).
Badro, D. A. et al. Y-chromosome and mtDNA genetics reveal significant contrasts in affinities of modern Middle Eastern populations with European and African populations. PloS ONE 8 (1), e54616 (2013).
Chiaroni, J. et al. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur. J. Hum. Genet. 18 (3), 348–353 (2010).
Platt, D. E. et al. Mapping post-glacial expansions: the peopling of Southwest Asia. Sci. Rep. 7 (1), 1–10 (2017).
Černý, V. et al. Out of Arabia—the settlement of island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity. Am. J. Phys. Anthropol. Off. Publ. Am. Assoc. Phys. Anthropol. 138 (4), 439–447 (2009).
Černý, V. et al. Regional differences in the distribution of the sub-Saharan, West Eurasian, and South Asian mtDNA lineages in Yemen. Am. J. Phys. Anthropol. Off. Publ. Am. Assoc. Phys. Anthropol. 136 (2), 128–137 (2008).
Černý, V., Čížková, M., Poloni, E. S., Al-Meeri, A. & Mulligan, C. J. Comprehensive view of the population history of A rabia as inferred by mt DNA variation. Am. J. Phys. Anthropol. 159 (4), 607–616 (2016).
Fernandes, V. et al. The Arabian cradle: mitochondrial relicts of the first steps along the southern route out of Africa. Am. J. Hum. Genet. 90 (2), 347–355 (2012).
Hodgson, J. A., Mulligan, C. J., Al-Meeri, A. & Raaum, R. L. Early back-to-Africa migration into the Horn of Africa. PLoS Genet. 10 (6), e1004393 (2014).
Bryc, K. et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. 107 (2), 786–791 (2010).
Plaster, C. A. Variation in Y Chromosome, Mitochondrial DNA and Labels of Identity on Ethiopia (UCL (University College London), 2011).
Luis, J. R. et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am. J. Hum. Genet. 74 (3), 532–544 (2004).
Reich D. [Available from: https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data.
Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536 (7617), 419–424 (2016).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513 (7518), 409–413 (2014).
Cavalli-Sforza, L. L. The human genome diversity project: past, present and future. Nat. Rev. Genet. 6 (4), 333–340 (2005).
Haber, M. et al. Insight into the genomic history of the Near East from whole-genome sequences and genotypes of Yemenis. bioRxiv 6, 749341 (2019).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81 (3), 559–575 (2007).
Patterson, N. et al. Ancient admixture in human history. Genetics 192 (3), 1065–1093 (2012).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19 (9), 1655–1664 (2009).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2 (12), e190 (2006).
Taleb N, Zalloua P, Elbassioni K, Henschel A, Platt D. Informational Rescaling of PCA Maps with Application to Genetic Distance (2023).
Willing, E. M., Dreyer, C. & van Oosterhout, C. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7 (8), e42649 (2012).
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461 (7263), 489–494 (2009).
Lawson, D. J., Hellenthal, G., Myers, S. & Falush, D. Inference of population structure using dense haplotype data. PLoS Genet. 8 (1), e1002453 (2012).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28 (19), 2520–2522 (2012).
Freed, D., Aldana, R., Weber, J. & Edwards, J. The Sentieon genomics tools - A fast and accurate solution to variant calling from next-generation sequence data. BioRxiv 89, 700 (2017).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38 (16), e164 (2010).
Bergström, A. et al. Insights into human genetic variation and population history from 929 diverse genomes. Science https://doi.org/10.1126/science.aay5012 (2020).
Almarri, M. A. et al. The genomic history of the Middle East. Cell 184 (18), 4612–25.e14 (2021).
Narasimhan, V. et al. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32 (11), 1749–1751 (2016).
Sankararaman, S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507 (7492), 354–357 (2014).
Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44 (W1), W58-63 (2016).
Chen, H., Lu, Y., Lu, D. & Xu, S. Y-LineageTracker: a high-throughput analysis framework for Y-chromosomal next-generation sequencing data. BMC Bioinform. 22 (1), 114 (2021).
Loh, P.-R. et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193 (4), 1233–1254 (2013).
Pickrell, J. K. et al. Ancient west Eurasian ancestry in Southern and Eastern Africa. Proc. Natl. Acad. Sci. U. S. A. 111 (7), 2632–2637 (2014).
Vyas, D. N., Al-Meeri, A. & Mulligan, C. J. Testing support for the northern and southern dispersal routes out of Africa: an analysis of Levantine and southern Arabian populations. Am. J. Phys. Anthropol. 164 (4), 736–749 (2017).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52 (W1), W78–W82 (2024).
Richards, M. et al. Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. Am. J. Hum. Genet. 72 (4), 1058–1064 (2003).
Kivisild, T. et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 75 (5), 752–770 (2004).
Rodriguez-Flores, J. L. et al. Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Res. 26 (2), 151–162 (2016).
Caton S. Yemen.: ABC-CLIO http://publisher.abc-clio.com/9781598849288 (2013).
Al-Jallad, A. The earliest stages of Arabic and its linguistic classification. In The Routledge Handbook of Arabic Linguistics (eds Benmamoun, Elabbas & Bassiouney, Reem) (Routledge, 2018).
Nebes, N. 299C53Early Saba and its neighbors. In The Oxford History of the Ancient Near East Volume V: The Age of Persia (eds Radner, K. et al.) (Oxford University Press, 2023).
Kitchen, A., Ehret, C., Assefa, S. & Mulligan, C. J. Bayesian phylogenetic analysis of Semitic languages identifies an early bronze age origin of Semitic in the Near East. Proc. Royal Soc. B Biol. Sci. 276 (1668), 2703–2710 (2009).
Acknowledgements
Computational work for the whole genome sequencing pipeline has been conducted on Khalifa University’s High Performance Computing center. Khalifa University’s Competitive Internal Research Award CIRA-2019-076, awarded to AH, was used to fund computational infrastructure. We thank the Wellcome Sanger Institute sequencing facility for generating data, supported by Wellcome grants 098051 and 206194. We would like to acknowledge the support of Professor Matthew Hurles for providing guidance and valuable comments that significantly strengthened the manuscript.
Author information
Authors and Affiliations
Contributions
A. H., C.A.H, E.P, S.A.K, and DP performed all the analyses. R. S.A., M.A.H and L.P., provided samples, worked on methods and provided background information and write-ups. A.H., N.N.T., D.P. and P.Z wrote the main manuscript and were behind the study design. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All subjects provided signed informed consent forms before participation in the study. The study was approved by the IRB (LAU.SOP.PZ1.2007.R4.6/November/2015) of the Lebanese American University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Henschel, A., Saif-Ali, R., Al-Habori, M. et al. Human migration from the Levant and Arabia into Yemen since Last Glacial Maximum. Sci Rep 14, 31704 (2024). https://doi.org/10.1038/s41598-024-81615-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-81615-4