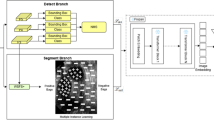
Abstract
Ovaries are of paramount importance in reproduction as they produce female gametes through a complex developmental process known as folliculogenesis. In the prospect of better understanding the mechanisms of folliculogenesis and of developing novel pharmacological approaches to control it, it is important to accurately and quantitatively assess the later stages of ovarian folliculogenesis (i.e. the formation of antral follicles and corpus lutea). Manual counting from histological sections is commonly employed to determine the number of these follicular structures, however it is a laborious and error prone task. In this work, we show the benefits of deep learning models for counting antral follicles and corpus lutea in ovarian histology sections. Here, we use various backbone architectures to build two one-stage object detection models, i.e. YOLO and RetinaNet. We employ transfer learning, early stopping, and data augmentation approaches to improve the generalizability of the object detectors. Furthermore, we use sampling strategy to mitigate the foreground-foreground class imbalance and focal loss to reduce the imbalance between the foreground-background classes. Our models were trained and validated using a dataset containing only 1000 images. With RetinaNet, we achieved a mean average precision of 83% whereas with YOLO of 75% on the testing dataset. Our results demonstrate that deep learning methods are useful to speed up the follicle counting process and improve accuracy by correcting manual counting errors.
Similar content being viewed by others
Introduction
Folliculogenesis is a highly complex and dynamic process which culminates with the ovulation of one or more oocyte(s) at each cycle. During each estrous cycle, the follicles develop from a dormant primordial pool. The oocytes start to grow and maturate while surrounded by an increasing number of granulosa cells. Various classifications have been used to describe the different stages of oocyte and follicle development1,2. Briefly, primordial follicles contain a partial or complete single layer of squamous granulosa cells. Primary follicles contain a single layer of cuboidal granulosa cells. Antral follicles are characterized by multiple layers of granulosa cells and a cavity named antrum. The remaining of the antral follicle following ovulation is called corpus luteum. It is composed of granulosa cells, thecal cells and blood vessels. The evaluation of follicle numbers across these different classes at various stages of development and/or upon exposure to hormonal/pharmacological treatments is crucial in many fields of biology. The number of follicles and corpus luteum can vary between estrus cycles in response to physiological and non-physiological factors. These factors include endocrine-disrupting chemicals3,4, maternal aging, chemotherapy5, infection6, and inflammation7. All of them have been shown to affect ovarian reserve. As the antral follicles and corpus lutea represent the hallmark of late follicular development and ovulation, counting their number is necessary when studying infertilities, improving assisted reproduction technologies or evaluating the effects of drugs.
Research and pre-clinical phase of drug development extensively use rodents as experimental models to evaluate potential efficacy and/or repro-toxicity8,9. Consequently, the refinement of follicle quantification methods has gained heightened significance. It is imperative for researchers to understand the strengths and weaknesses of the available counting approaches to ensure the accurate interpretation of results. Histological counting have been widely accepted and used in reproductive biology research to estimate the number of follicles. It enables the distinction of various types of follicles including, primordial, primary, secondary, and antral follicles. It offers spatial distribution and organization of follicles within the ovary. Follicle detection by traditional methods, such as manual counting or semi-automated techniques10, often face limitations in terms of accuracy, efficiency, and consistency. More recent studies have used semi-automated techniques (e.g., region growing, active contours, outer or inner follicle boundary) to assist in follicle detection, but these still suffer from issues related to false positives, poor generalization across different imaging conditions, and computational inefficiency. The semi-automated techniques still require human intervention in refining and validating the detection results. While this can be efficient in some cases, it can also be time-consuming and prone to human error, especially in larger datasets or more complicated cases.
With the rise of machine learning and deep learning technologies in many domains such as image recognition11, robotics12, speech recognition13, life sciences14,15,16,17, etc, there has been a shift towards automating the detection of ovarian follicles. This shift aims to improve the accuracy, efficiency, and consistency of diagnostic workflows, which are often hindered by the manual and time-consuming nature of traditional methods. AI models have shown great performance in image analysis due to the availability of large amount of labelled dataset, sometimes even surpassing the contributions of experts18. Most of the existing deep learning-based models in medical image analysis typically focus on image classification or segmentation or object detection. In image classification, the model would simply tell you whether follicles are present in the image, but it would not tell you how many follicles or where they are located in the image. This would be helpful in scenarios where the task is to quickly determine whether a follicle is visible in an ultrasound scan, but it is a major limitation for tasks that require precise localization and counting. Object detection, on the other hand, goes a step further than image classification by not only identifying what is in an image, but also localizing the objects of interest. In the case of follicle detection, the goal is to not only identify and count follicles, but also to locate their positions within the image.
Follicle detection from histology images using deep learning methods remains largely an uncharted territory. The high resolution of whole slide digital images (WSI) obtained from digital slide scanners, combined with advanced AI methods, can reduce the workload and inconsistencies of current approaches19,20. This paper highlights the benefits of AI techniques, particularly deep learning, for counting antral follicles and corpus lutea. Our work is based on object detection techniques to localize and count structures of interest. Object detection models can be trained on diverse datasets and are robust against various conditions such as differing image qualities, noise, or follicle shapes. We utilized object detection models, including YOLO (You Only Look Once) and RetinaNet, to localize objects and improve detection accuracy.
Regarding previous work on deep learning methods for counting follicles in mouse ovaries, Sonigo et al.21 proposed a convolutional neural network (CNN) with a sliding window algorithm to count primordial follicles. They used a dataset comprising 9 million images of mouse ovaries for training and 3 million images for testing. The model achieved a precision of 65% and a recall of 91%. These values were obtained after applying hard negative mining and conducting manual checks to reduce the large number of false positives. Compared to21, which developed a two-step approach for follicle detection, our method is based on an object detection approach that provides localization by generating bounding boxes around individual follicles and predicting their classes. While their work focused on a single follicle type, our approach detects and differentiates between two types of follicles. In a later study, Inik et al.22 performed the detection of five classes of follicles: primordial, primary, preantral, secondary, and tertiary. They began by generating sub-images from the input image, classifying these sub-images into edge, follicle, and background classes. Finally, they created a binary image representing the background and follicle classes, overlaying these binary sub-images onto the original input image. In the final classification phase, all follicles localized in the input image were classified into the five classes. Their dataset consisted of 1,750 images for training and 222 images for testing. On the testing dataset, they achieved a mean accuracy of 95%. In contrast to22, who used CNN-based segmentation, our method allows for the detection of individual follicles with localization information using object detection techniques. Additionally, the real-time detection capability of YOLO makes our method more scalable and faster than traditional approaches, which is essential for clinical environments requiring high-throughput analysis. Unlike earlier studies, which detected all follicle types (primordial, primary, preantral, secondary, and tertiary), our study focuses on late-stage follicles, specifically antral follicles and the corpus luteum. Furthermore, we report state-of-the-art mean average precision (MaP) metrics for evaluating the proposed object detection models, which were absent in the aforementioned studies. The proposed model represents a first step towards automating the quantitative assessment of late folliculogenesis.
The remainder of the paper is structured as follows. In Section 2, we introduce the background on late follicles, their annotation, and describe the proposed machine learning framework for follicles counting. In Section 3, we present and compare our results. In Section 4, we identify limitations, give concluding remarks and describe future work.
Methods
Animals
All experimental and care procedures were carried out in strict accordance with the relevant guidelines and regulations of the European and French Directives. Experiments were approved by the local Ethical Committee of Animal Experimentation CEEA Val de Loire \(\hbox {N}^\circ\)19 and the French ministry of teaching, research, and innovation (APAFIS #18035-2018120518194796). C57BL/6JOlaHsd mice were purchased from Inotiv.inc. The 10 female mice of 12-20-week-old were conventionally housed in groups of 4 in type 2 cages in the rodent animal facility, experimental unit: UEPAO (PAO, INRAE: Animal Physiology Facility, https://doi.org/10.15454/1.5573896321728955E12) in an environmentally controlled room maintained at \(21^\circ\)C, humidity of 55% with a 12h light - 12h dark photoperiod, ad libitum access to food and water. Mice were allowed to acclimate within the UEPAO for at least one week prior to any procedures.
Tissue collecting and processing
All experiments were performed on animal materials postmortem. Results are reported in accordance with the relevant points of the ARRIVE guidelines. The mice were euthanized by cervical dislocation. Ovaries collection was performed between 9:00 and 11:00. They were trimmed from the fat pad and fixed in Bouin’s solution (Sigma Aldrich, HT10132) at \(4^\circ\)C overnight. The samples were dehydrated using ethanol-water sequential incubations and embedded in paraffin blocks (see 1). They were sequentially sectioned into 7 \(\mu\)m using a microtome (Leica HistoCore AUTOCUT). The whole sections were mounted on microscope Superfrost Plus slides. Between 7 and 15 consecutive sections were placed onto a single slide. After 48h at room temperature, each slide was deparaffinized, rehydrated, and stained with hematoxylin-eosin (Sigma Aldrich, HHS32, HT110132). The sections were mounted in Depex (DPX new, Merck GaA, Darmstadt, Germany).
Manual follicle counting
The slides were digitized after 72h using a histology slide scanner Axio scan Z.1 Zeiss, running under Zen software (ZENblue 3.5 edition) with a magnifcation of 10x (numerical aperture 0.45) (see Fig. 1). Follicles containing multiple layers of granulosa cells and a follicular antrum were designed as antral follicles. To avoid counting the same follicle on serial sections, only those containing a clear visible oocyte were scored. The total number of antral follicles is the sum of the antral follicles from all sections of a complete ovary. The corpus luteum is more a solid structure, made of granulosa cells (rounded cells), theca cells (elongated cells), and blood vessels.
Deep learning framework for automatic counting ovarian follicles
CVAT annotation and data extraction
The sections on the slides were extracted in Joint Photographic Experts Group format (JPG) by using ZENblue 3.5 edition to annotate with Computer Vision Annotation Tool (CVAT), a free open source, suitable for image and video labeling23. We decided to use bounding boxes for annotation purposes for two reasons: (1) Boxes require relatively less workload to annotate as compared to other formats such as polygon, etc. (2) It has been shown previously that other formats do not necessarily increase performance24 by large margin while causing more workload for the annotator. Three structures were annotated: (1) antral follicle (AF), (2) antral follicle without ovocyte (AFWO), and 3) corpus luteum (CL). In Fig. 1, we show the manual annotation workflow.
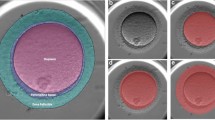
Data extraction process for annotation of whole slide images. (a) Sample processing - Ovaries were dissected out and trimmed from the fat pad, fixed in Bouin’s solution, dehydrated and embedded in paraffine blocks. (b) Image digitation - The stained slides were digitized by Axio scan Z.1 Zeiss with a magnification of 10x. (c) Follicle annotation - Follicles containing multiple layers of granulosa cells and a follicular antrum were designed as antral follicles (Pink box), The antral follicles lacking a visible nucleus were labelled as antral follicle without oocytes (Blue box) and the temporary endocrine structures formed from the remanants of the ovarian follicle after ovulation were identified as corpora luteum and annotated (Yellow box).
Data augmentation
Data augmentation entails creating the augmented or fake dataset to increase the size of the training dataset which helps to improve the performance of deep learning models and to tackle the issue of overfitting. For some kind of datasets it is fairly easy to perform augmentation, for example on imaging dataset, one can perform basic image transformations such as like rotation, resizing, and translation by few pixels25. We can divide the data augmentation into online and offline methods depending on when the augmentation is being performed. Online augmentation is a real-time process in which images are enhanced on-the-fly during model training, whereas offline augmentation requires images to be modified ahead of time, subsequently added into the dataset, and then later loaded into the memory for training. In our work, we use on-the-fly data augmentation since it eliminates the need to save additional datasets. This approach not only conserves storage space but also enhances computational efficiency by dynamically generating augmented data during the training process, thereby improving model generalization and robustness.
Class imbalance
Classification algorithms are known to be very sensitive to unbalanced data when the aim is to derive classification and prediction tools for categorical classes. In general, the algorithms will correctly classify the most frequent classes and lead to higher misclassification rates for the minority classes, which are often the most interesting ones. In our case, we observe that the AFWO is an over-represented class. We can see from Table 1 that we have approximately 1.5x more samples of AFWO as compared to AF and approximately 2x more samples of AFWO as compared to CL. To deal with the data imbalance, we sampled the dataset as shown in the sampled dataset column. In object detection tasks, we have foreground-background class imbalance in addition to foreground-foreground class imbalance. It is unavoidable because majority of the boxes are labeled as the background during the training process. In this paper, we employ sampling methods to deal with the foreground-foreground imblance. Refer to26 for a detailed review of various data imbalance strategies.
Object detection models
Object detection includes both locating and classifying objects of interest in images (in our case, full sections of ovaries). In general, two-stage and one-stage detector models are used in object detection. Two-stage detectors are slower but more accurate due to their sequential approach to objects detection and classification. A one-stage detector locates and classifies objects in parallel, making it faster but slightly less accurate than two-stage detectors. In our work, we compare the performance of two one-stage detectors: RetinaNet27 and YOLO28. Since our objective is to choose an architecture with real-time detection capabilities, we have opted to utilize one-stage detectors. We adopt the transfer learning approach to improve performance of our models by utilizing pre-trained models on other tasks29. Additionally, we utilize the early stopping criteria to halt the training of a model when its performance no longer improves.
In Fig. 2, we show high level overview of RetinaNet and YOLO (You Only Look Once). Detailed architectures can be found in supplementary files (S1 and S2). RetinaNet is a popular one-stage detector which combines feature pyramid network (FPN) for detecting multi scale objects and focal loss to handle imbalance between background and foreground class. RetinaNet has two separate output detection head, one for the classification and one for the bounidng box regression. These heads are shared among all the features of the FPN. We used different backbone (ResNet50, CSPDarkNet, MobileNetV3, EfficientNetV2) architecture trained previously on ImageNet 2012 to gauge difference in performance and to establish baseline performance. Our goal is to select a backbone model which can be used to build a tool used by biologists with real-time performance.
YOLO is extremely fast, real-time one-stage detector where a single neural network is used to simultaneously predict multiple boxes and their classification probabilities. YOLO is relatively less accurate but incredibly fast object detection architecture, it is commonly utilized in security cameras. YOLO divides an entire image into a S x S grid, and if the center of a certain object falls within the grid cell, then this grid is responsible for detecting that specific object. Each grid cell can predict several bounding boxes along with their confidence scores and classes. The confidence score indicates whether or not the object has been detected. YOLO multiplies the class probabilities for each grid cell with confidence scores of the bounding box to obtain final detections. In our work, we use the most latest variation of YOLO called YOLOv8 with a small backbone YOLOV8 pretrained on COCO dataset. This architecture comes with two major modification, i.e., anchor-free detection and mosaic data augmentation.
High level overview of one-stage detectors. RetinaNet employ feature pyramid network to extract multiple scales information. Detection heads takes information from multiple levels to perform prediction. YOLO takes the whole image, resize it, apply single convolutional network over it, and finally employs non-max suppression based on model’s confidence scores.
Evaluation metrics
In this paper, we use state-of-the-art metrics to evaluate the performance of our object detection models, i.e., average precision (AP). AP relies on precision, recall, and intersection over union (IoU) metrics.
Intersection over union
In object detection, we localize objects and predict their classes using boundary boxes. Object detection models take into account the quality of the predictions by calculating the intersection between the ground truth object box G and the predicted bounding box \(\overline{\textrm{G}}\). IoU represents the ratio of the intersection over union between the ground truth and predicted boxes (see Eq. 1). We generally measure the performance of the object detection model using various IoU thresholds. In Fig. 3, we highlight different IoU thresholds, orange represents the predicted box while blue represents the ground truth box.

where A represents the area of the bounding box.
Precision and recall
The term “precision” is used to describe how accurately our model identifies correct predictions, i.e., how many of the total detections for a given class actually belonged to that class. Recall refers to the proportion of instances of a particular class that our model correctly predicts out of the total ground truth instances for that class. There is typically a trade-off between precision and recall; increasing one often result in a decrease in the other. We aim to increase the precision and recall values as much as possible. The precision and recall values are calculated using Eqs. (2 and 3), respectively.
A true positive (TP) represents the number of true predicted boxes where IoU is equal to or greater than a certain threshold. A false positive (FP) is a predicted bounding box that either does not match any ground truth box (IoU is below the threshold), incorrectly matches the class label, or is an extra detection when multiple boxes are predicted for the same object (only one is kept as TP). A false negative (FN) indicates the model’s inability to identify an object present within the image, i.e., no predicted box overlaps with the ground truth box above the IoU threshold or the predicted box overlaps but the class is not correctly identified.
Average precision
Average precision (AP) can be used to summarize the precision and recall values into a scaler. It represent the area under the precision-recall curve and is calculated using Eq. (4) for each class:
where p is the precision and r is the recall.
We calculate mean average precision (MaP) using using Eq. (5):
where N is the total number of classes, in our case 3, \(AP_i\) is the average precision for class i, and MaP is the average precision of all class’s average precision.
Results
Image annotations
Since we are performing supervised object detection task, it is necessary to obtain labeling data of objects in different histology images. We use CVAT to annotate images to identify category of follicles and their boundary information. In Fig. 4, we show the annotation of one whole slide image in the CVAT software. Approximately 60 hours were required to annotate the whole data set. A total of 1373 antral follicles with nucleus, 1941 antral follicle without nucleus and 869 corpus luteum.
Annotation of whole slide images (WSI) using the Computer Vision Annotation Tool (CVAT). This figure illustrates the CVAT interface used for the annotation of ovarian structures for image analysis. The interface displays an image of an ovary section with annotated regions highlited by colored boxes. The yellow box indicated the area corresponding to the corpus luteum. The blue box delineates an antral follicle lacking a visible nucleus. The pink box represented an antral follicle with a discernible nucleus.
The input data in our model consists of four numeric values which represent where the object is located on the image and one more value that shows what kind of an object it is. Different formats can be used to represent boundary boxes. For example, one can define the boundary box by specifying a set of corner coordinates, width and height. Another way is to describe it using the coordinates of its top-left and bottom-right corners, i.e., \([x_1,y_1,x_2,y_2]\). To specify coordinates in our study, we use the [x, y, w, h] format. The top-left corner is represented by two values x and y, width and height are represented by the values w and h.
Data description
Our full dataset consists of 1209 ovarian sections. Within these sections, we counted 1373 antral follicles, 1941 antral follicles without oocytes, and 869 corpus lutea. The sampled dataset consists of 999 images with 1373 antral follicles, 1549 antral follicles without oocytes, and 869 corpus lutea objects. Both datasets were randomly divided into training (70%), validation (15%) and testing (15%) sets. In Table 1, we show characteristics of our dataset.
Preprocessing
We converted XML files obtained through the CVAT software to CSV files to perform object detection tasks. We use the OpenCV python library30 to remove the excessive white patches from histology images.
It is well established that image resolution has a direct impact on the classification and localization of objects. In particular, it is difficult to detect small objects in low-resolution images. In general, for CNN-based detectors, images are down-sampled to a resolution of 256 x 256. Our initial findings confirmed that resolution does impact the performance of object detectors. We observed that high resolution, although computationally more expensive, does not translate systematically into improved performance, which has also been shown previously in other studies31,32. In our case, we set the image resolution to 640 x 640 pixels. The quantitative analysis of resolution impact on computational time and accuracy of the model can be found in the supplementary files (S5 and S6).
To deal with the background-foreground class imbalance, we used focal loss27. This loss function modifies classic cross entropy function in a way that it down weights the loss contribution of well classified examples (background class) and quickly leading the model to focus on difficult examples. In order to deal with the object class imbalance, we used the data sampling technique. We kept only those images that had at least one of the antral follicle or corpus luteum objects, leading to the elimination of 210 images. Note that these images did not have any antral folllicle and corpus luteum objects. In this paper, we refer to this dataset as a sampled dataset.
Data augmentation in object detection models is more challenging and complex as compared to simple classification models as we must take into account the underlying bounding boxes during various transformations. We performed on-the-fly data augmentation using Keras-CV library33, which propose native support for data augmentation with bounding boxes. We used random flip and jittered resize techniques for increasing the diversity of our dataset. In Fig. 5, we show two images from our training set before (see Fig. 5a) and after augmentation (see Fig. 5b).
Comparative models
The experiments were performed on a server with a NVIDIA A30 24GB PCIe NonCEC Accelerator GPU card. We build our dataset using tensor flow data API and models using keras-CV library33.
The dataset is divided into training, validation and testing sets. During the training phase, the model parameters (weights and biases) are updated. During model training, the validation of the model is carried out by calculating the MaP using the validation dataset. The model with the highest MaP is saved, and it is subsequently used to make predictions on the testing dataset. We employed a stochastic gradient descent (SDG) optimizer with a batch size of 16. Exploding gradient is a common problem that arises when developing object detection models. When we train neural networks, an error gradient captures the magnitude and direction of the updates we make to the weights in a particular direction by some amount. The huge update to the model parameters caused by the high gradient values results in an unstable network. Gradient clipping, which relies on a threshold value, is used to cope with the exploding gradient problem. When a gradient value exceeds a predetermined threshold, the value is set to the threshold’s value. After completion of the training phase, we use testing dataset to measure the model’s generalizability. In Fig. 6, we show loss function of the object detectors, orange represents the validation loss and blue represents the training loss. We observe that there is a sudden change in gradient in the beginning of the training which stabilizes with time.
Graphs of loss function. It illustrates the training and validation loss curves for YOLOv8 and RetinaNet models on the employed dataset. The x-axis represents the number of epochs during the training process. The y-axis represents the value of the loss function. The goal during training is to minimize this value, as a lower loss indicates better alignment between the model’s predictions and the actual data.
In Table 2, we show comparison of full dataset and sampled dataset using RetinaNet detector. It takes \(\approx ~22h\) to train on a full dataset without any performance gain as can be observed from Table 2, while it takes approximately \(\approx ~12h\) hours of training on sampled dataset. In addition to the computational overhead, model trained on the full dataset does not performed well on CL class as compared to the WO and AF class using validation and testing sets (IoU 0.50). Furthermore, our model built using sampled dataset is able to classify the remaining 200 images with a MaP of 0.83 (IoU 0.50) which were initially deleted from the dataset to create a more balanced dataset. Finally, it is important to note that we did not experience a loss in performance for the AFWO class (the overrepresented class); rather, performance improved in all cases, except for the IOU of 0.75 in the 6th row of Table 2. We include the results with two different IoU threshold, i.e., 0.50 and 0.75. The comparison based on different IoU threshold can be found in the supplementary file (S4). It is worth noting that different thresholds can lead to some side effects. For example in case of high threshold, we may filter or eliminate predicted boxes for overlapping objects, which is not a desirable behavior. There is no ideal IoU threshold value, it varies between 0 and 1. In practice, the common IoU threshold is 0.5 and may need customization in a particular context to obtain meaningful outcomes. Note that our model obtained recall of 0.61, precision of 0.83, and F1 score of 0.71 on the testing dataset. A detailed comparison of these metrics for each class on training, validation, and testing sets can be found in the supplementary file (S3).
As a sanity check, we also trained the model without any data augmentation to ensure the robustness of our model. The initial results of this experiment indicate an overfitting problem; as our model is able to achieve MaP of 0.99 (IoU 0.50) on the training dataset However, these impressive results raise concerns about generalizability of the model, i.e., predictions quality on the unseen dataset. Taking into account this concern, we applied on-the-fly data augmentation techniques, which effectively improved the model’s generalizability on the unseen dataset. Furthermore, to optimize the computational resources, we converted images into grey-scale. Our results did not show any significant performance gains in the context of our dataset and model.
In Table 3, we show comparison of YOLO and RetinaNet detectors with different backbone architectures. When it comes to compution speed, YOLO outperforms RetinaNet by a wide margin. It requires roughly only one hour to complete the training. RetinaNet with MobileNet as backbone surpassed YOLO on the testing dataset with a MaP of 0.86. As compared to RetinaNet, the confidence threshold for predicted bounding boxes was quite low. Additionally, YOLO achieved a MaP of 0.71 (IoU 0.50) on the remaining dataset (absent from the training, testing and validation sets) while RetinaNet scored a MaP of 0.83 (IoU 0.50). We believe that the RetinaNet is a suitable choice where computational time is not an issue and the top priority is MaP. However, where we can tolerate slightly lower MaP for the sake of faster performing algorithm, a YOLO model is more suitable option.
In Fig. 7, we show the results on the testing dataset. In blue are the original annotations and in yellow are the predictions obtained through RetinaNet with MobileNetV3 as backbone. We are using the batch size of 8, IoU threshold and confidence score of 0.75. We observe that our model failed to predict two objects in Fig. 7b and one in Fig. 7c. Figure 7d displays an antral follicle lacking a nucleus, intentionally left unannotated by the expert but correctly labeled by the detector. Despite being located within a distorted region, the follicle was accurately detected. This distortion is attributed to artifacts arising from slide mounting. Although the artifact affects the region, it does not significantly alter the structure’s shape, allowing for the follicle’s identification through the predictions. Figure 7f shows an antral follicle that was forgotten by the annotator but detected correctly by the trained model. These were considered as FPs in our evaluation criteria, which means we penalized our model for detecting these follicles. These detections highlight the labeling errors. In future research, we plan to enrich the dataset by correcting these errors which will help to further optimize the training of our models and to create a more general model.
In Fig. 8, we show the counting by expert and predictions by our model on the testing dataset with IoU threshold of 0.75 and 0.50. The blue color represents the follicle counting performed by the expert, orange represents the predictions by the model, pink signifies the TPs, and green shows the FPs. We see that the TPs rate increases and FPs rate decreases with the most common IoU threshold of 0.50. This figure highlights the effects of IoU threshold which requires careful consideration for different applications.
Furthermore, we analyzed images manually to understand logic behind the predictions of our models, which is crucial for further studies to understand why errors occur and how to address them. This comparative analysis of expert and model identification helps to elucidate the strengths and limitations of AI-driven image classification in the context of ovarian tissue analysis. We examined 151 images manually from the testing set to identify where the model misclassified, confused, or failed to label follicles correctly. What we found particularly intriguing and promising was the agreement between the number of false positives predicted by the model and those by the operator, especially for the antral follicle and the corpus luteum, the two main important structures for us. Upon closer examination, we observed that not all discrepancies were genuine errors made by the model (see Fig. 9).
Errors analysis of expert and model identification of ovarian structures. This figure presents section of ovarian tissue with structures identified by both human expert (Blue bounding boxes) and AI model (Yellow bounding boxes). The expert categorized discrepancies into three types of errors: (1) The geniune or real errors (Red boxes) : structures identified by experts but missed by the model. These are clear structures that the model should have recognized but failed to do so, (2) Allowable erros (Light red boxes) : structures recognized by the expert but not labeled by the model due to staining inconsistencies, mounting issues, scanning artefacts, or other technical factors, and (3) Errors corrected by the model (Green Boxes) : structures overlooked by the expert due to factors such as eye fatigue or intentionally unlabelled (tears in the sections) but correctly identified by the AI model. These instances higlight the model’s ability to detect structures that may have been missed or intentially omitted by the human expert.
Some regions posed challenges for labeling due to staining inconsistencies, mounting issues, and scanning artifacts, making them difficult even for the operator to label accurately. We categorized these instances as allowable errors given the challenges in image acquisition and processing. Despite these challenges, the system demonstrated an ability to detect and correctly classify some follicles that were missed by the experimenter. Thereby, after reclassification of these errors into those corrected by the model, those allowable by the system, and genuine/real errors, we noted a reduction in the number of false positives (see Fig. 10). This is expected to significantly enhance precision and consequently improve the accuracy of the model.
Comparative analysis of False positives (FP) detected by the model and manually by the operator before and after analysis of 151 images from the testing set. The light green bars represent the number of FP identified by the model with a precision of 0.75. The green bars with diagonal lines (OFP-Before analysis) depict the number of FP manually counted by the operator upon initial comparison of the images. The green bars with vertical lines show the number of FP (OFP- After analysis) counted by the operator after reclassification of errors between Geniune, allowable and corrected errors. The decrease in the number of FP observed after manual examination and eror correction by the operator demonstrates the potential of enhanced precision and accuracy of the model.
Discussion
In this paper, we present a valuable technique to count ovarian follicle from whole slide images (WSI) using transfer learning, early stopping and data augmentation approaches. We annotated the WSI of 20 mice ovaries in CVAT. From these annotations, a model trained; validated and tested itself to distinguish between three different classes of follicles: the antral follicle, the antral follicle without nucleus, and the corpus luteum. We used state-of-the-art one-stage object detection methods, i.e, YOLO and RetinaNet for follicles detection in histology images. We achieved a MaP of 0.83 on the testing dataset with RetinaNet. Furthermore, we identified cases where model was able to correct erros of the annotators.
Histological counting may be one of the most standardized approaches for assessing ovarian reserve and follicle status, it does come with certain limitations: (1) Subjectivity of the operator : even with the presence of skilled personnel, there is a potential inter-observer variability due to the manual nature of histological counting. This variability may lead to inconsistencies in follicle counts, (2) Time-consuming and tedious work: the histological process requires careful preparation, sectioning, staining and examination. Each step in the preparation may affect the accuracy and quality of follicle counting. Studies show that we should take in consideration the impacts of different fixatives (Bouin’s fixative versus formalin), embedding material and section thickness34. They do not have the same effects on tissue structure. Despite the common use of formalin, Bouin’s solution may preserve the cellular morphology better than formalin that may cause tissue shrinkage and consequently changes in follicle dimensions. Results may not be fully representative of the entire ovary if sections at a regular interval were analyzed. Correction factors were used in such cases19. In our case, we used Bouin’s solution to fix the collected ovaries, a standardized hematoxylin and eosin staining protocol that selectively highlight the different aspects of tissue. The entire ovary was cut and almost all the slides were digitized. There was not any specific selection of the slides which reduce the introduction of sample bias. The use of a scanner helped to digitize faster and with high resolution the Whole Slide Images. The contrast quality of the images improved the ability to analyze. Despite the efforts, staining inconsistency, mounting or scanning artefacts may continue to exist. Hence, some structures were intentionally unlabelled by the experiment especially on slides with mounting or scanning artefact. This was not a problem for the model, we noticed that the model was able to predict correctly the follicles in some poorly prepared sections with distorted aspect. Some experimenter labeling errors were found demonstrating that fatigue can contribute to errors and were discovered when comparing results obtained through the automated method with manual histological labeling. The scientists need to be always on a focused state for accurate results. One more reason to have at least two experimenters to assess follicle counting. Using AI and mainly this model can indeed reduce the need for multiple experimenters. The model labeled correctly almost all the structures but it only missed some blur structures (mounting or scanning artefacts) that the experimenter was able to annotate when checking the successive slide.
Taking in consideration these limitations is crucial for scientists when designing and interpreting results. Integrating histological counting with additional methodologies like hormone dosage and embracing new technologies, such as artificial intelligence and deep learning can help as we can see to overcome some of the challenges of ovarian follicle counting. Histological counting is often used as a benchmark for validating automated techniques, including those involving artificial intelligence. Comparing results obtained through automated methods with manual histological counting helps ensure the accuracy and reliability of the automated approach. Understanding the nature of errors and discrepancies is crucial for refinning AI algortithms and optimizing their performance in biomedical imaging applications. The model shown in this paper, offers solutions to enhance accuracy, efficiency, and reproducibility of follicle counting, when comparing the predicted labelling and the annotated one.
The approach used in the paper is applied to the mouse ovary but can be extrapolated to ovaries of other species, including humans by transfer learning. It is already planned to use proposed method on histological slides of sheep and rabbits. In fact, the mouse ovary is considered as a valuable model. It allows to perform various genetic modifications, such as gene knockouts and knock-ins. By selectively disabling or altering specific genes, scientists can gain insights into the molecular pathways that regulate follicle growth and health. These modifications enable detailed studies of folliculogenesis. Number of follicles per ovary can be counted then in both healthy and diseased ovaries such as in PolyCystic Ovary syndrome (PCOs). Moreover, Deep learning is transforming how pathologists analyze histological slides, making their work both more efficient and precise. For instance, research by35 demonstrated that all slides featuring prostate cancer, along with micro- and macro-metastases from breast cancer, could be identified automatically. Remarkably, this process allowed for the exclusion of \(30-40\)% of slides containing benign or normal tissue, all without the need for additional immunohistochemical markers or human intervention. Additionally,36 and37 applied deep learning to analyze WSIs in lung and brain cancers, revealing previously unidentified patterns and contributing to the development of highly accurate prognostic models. Similarly,38 and39 harnessed deep learning to analyze whole slide images (WSIs) for colorectal cancer prognosis, focusing on various aspects such as cancer cell structure and tissue morphology. Their efforts resulted in the creation of more precise predictive models for cancer prognosis and progression. For liver cancer40, utilized deep learning models to uncover intricate patterns in WSIs, enhancing our understanding of the disease’s progression. In another study41, employed deep learning to analyze WSIs and identify specific morphological patterns in breast cancer tissues. Their research has significantly contributed to developing highly accurate prognostic models, showcasing deep learning’s ability to manage complex and high-dimensional WSIs effectively42. also explored the use of deep learning with WSIs in bladder cancer prognosis, highlighting the potential for tailoring more personalized treatment approaches. Our model can process vast amounts of imaging data quickly, allowing for rapid detection and the ability to check morphological changes that might be overlooked by the human eye. By leveraging advanced neural networks, deep learning algorithms can automatically identify and classify complex patterns within tissue samples, often catching subtle changes that might go unnoticed by the human eye. This technology helps pathologists by highlighting areas of interest and prioritizing cases, which can be especially valuable in busy clinical settings. With deep learning, pathologists can focus more on interpreting results and making informed decisions, rather than getting bogged down in routine analyses. Ultimately, this collaboration between technology and human expertise not only streamlines the diagnostic process but also enhances the quality of patient care, paving the way for more personalized treatment options.
There are some limitations of this study: (1) the experimental dataset is collected from the same laboratory which means we may not have enough diverse images in our training dataset, (2) optimal threshold to count the true or false positives remains dependent on the application, and (3) black-box nature of the deep learning models which means the thought process behind a particular decision or prediction of DL models is humanly non-interpretable due to complex non-linear internal structure and over parametrization.
In the near future, we want to create a larger and more diverse dataset by collaborating with other labs to create a more general detection models and to futher improve the MaP score. By inspecting the results manually, as discussed above, our model was able to produce predictions where annotator failed to label. In the current model, we penalize it for detecting these structure. In future, we will correct these errors and based on these intuitions, we will perform experiments to define the optimal threshold for each class separately to count the true number of true or false positives.
In biological contexts, understanding the internal workings of deep learning models is crucial, especially when trying to identify the most essential features or reasons behind classification or regression tasks. Over the past decade, numerous methods have been developed to tackle the problem of the explainability of deep learning models, such as feature relevance, local or global explanations, and visualizations [for a review, see 43,44]. However, these approaches are not always directly translatable to object detection tasks, which involve multiple outputs, such as bounding boxes, object categories, and their locations within the image. In the case of classification tasks, our output is normally scalar; however, in the case of object detection, the output is multiple bounding boxes per image, i.e., the bounding box coordinates of the detected objects and their category. The task is further complicated because of non-max suppression which is applied to eliminate overlapping boxes and retain only one box per detected object. It implies that we cannot translate the input to the output via the usual gradient, which hinders our ability to technically apply methods such as DeepLift, ShAP, etc. Moreover, explanations in object detection models not just needed for the category but also for the location of the objects. Generally, there has been a limited effort to tailor explanation methods for object detection models 45,46,47,48. These approaches cannot systematically be applied to our models because these methods often depend on the specific characteristics of the object detectors, such as one-stage or two-stage detectors, presence of anchor boxes 45 (our YOLO-based model is anchor-free), absence of individual explanations for the object category and bounding box 48. The method proposed in 46 is the model agnostic to generate heatmaps for highlighting the important parts of the image, however it is computationally slow and lacks evaluation of the explanation of bounding boxes. While these methods contribute valuable insights, further development is necessary to bridge the gap in the development of robust explainability tools tailored to object detection. In future, we will work on explainability of our object detectors to identify the causes of its predictions for model validation and knowledge discovery. For example, in order to improve computational overhead, we can think of explaining the prediction at the layer level rather than at the image level. Another future direction could be to work on the quality of the explanation for particular locations by using or developing quantitative metrics. Furthermore, we can think of adding domain knowledge (such as shape or size) to emphasize the biologically meaningful part of the image. These techniques will not only improve model interpretability but also allow for a comparison between the expert annotations and model intuitions for model validation and discovery of knowledge. We believe that these steps will further improve the performance and usability of our models.
Data availability
The data that support the findings of this study are available from the corresponding author.
Code availability
The scripts are available at the following DOI: 10.5281/zenodo.14192196.
References
Pedersen, T. & Peters, H. Proposal for a classification of oocytes and follicles in the mouse ovary. Reproduction 17, 555–557 (1968).
Hadek, R. The structure of the mammalian egg. Int. Rev. Cytol. 18, 29–71 (1965).
Ge, W., Li, L., Dyce, P. W., De Felici, M. & Shen, W. Establishment and depletion of the ovarian reserve: physiology and impact of environmental chemicals. Cell. Mol. Life Sci. 76, 1729–1746 (2019).
Panagiotou, E. M., Ojasalo, V. & Damdimopoulou, P. Phthalates, ovarian function and fertility in adulthood. Best Practice Res. Clin. Endocrinol. Metabol. 35, 101552 (2021).
Morgan, S., Anderson, R., Gourley, C., Wallace, W. & Spears, N. How do chemotherapeutic agents damage the ovary?. Hum. Reprod. Update 18, 525–535 (2012).
Santulli, P. et al. Decreased ovarian reserve in hiv-infected women. AIDS 30, 1083–1088 (2016).
Cui, L. et al. Chronic pelvic inflammation diminished ovarian reserve as indicated by serum anti mülerrian hormone. PLoS One 11, e0156130 (2016).
Winship, A. L., Bakai, M., Sarma, U., Liew, S. H. & Hutt, K. J. Dacarbazine depletes the ovarian reserve in mice and depletion is enhanced with age. Sci. Rep. 8, 6516 (2018).
Winship, A. L. et al. The parp inhibitor, olaparib, depletes the ovarian reserve in mice: implications for fertility preservation. Hum. Reprod. 35, 1864–1874 (2020).
Sarty, G. E., Liang, W., Sonka, M. & Pierson, R. A. Semiautomated segmentation of ovarian follicular ultrasound images using a knowledge-based algorithm. Ultrasound Med. Biol. 24, 27–42 (1998).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Károly, A. I., Galambos, P., Kuti, J. & Rudas, I. J. Deep learning in robotics: Survey on model structures and training strategies. IEEE Transactions on Systems, Man, and Cybernetics: Systems (2020).
Graves, A., Mohamed, A.-r. & Hinton, G. Speech recognition with deep recurrent neural networks. In: 2013 IEEE international conference on acoustics, speech and signal processing, 6645–6649 (IEEE, 2013).
Ching, T. et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018).
Min, S., Lee, B. & Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 18, 851–869 (2017).
Razzak, M. I., Naz, S. & Zaib, A. Deep learning for medical image processing: Overview, challenges and the future. In Classification in BioApps, 323–350 (Springer, 2018).
Razzaq, M. et al. An artificial neural network approach integrating plasma proteomics and genetic data identifies plxna4 as a new susceptibility locus for pulmonary embolism. Sci. Rep. 11, 14015 (2021).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Tilly, J. L. Ovarian follicle counts-not as simple as 1, 2, 3. Reprod. Biol. Endocrinol. 1, 1–4 (2003).
Myers, M., Britt, K. L., Wreford, N. G. M., Ebling, F. J. & Kerr, J. B. Methods for quantifying follicular numbers within the mouse ovary. Reproduction 127, 569–580 (2004).
Sonigo, C. et al. High-throughput ovarian follicle counting by an innovative deep learning approach. Sci. Rep. 8, 13499 (2018).
İnik, Ö., Ceyhan, A., Balcıoğlu, E. & Ülker, E. A new method for automatic counting of ovarian follicles on whole slide histological images based on convolutional neural network. Comput. Biol. Med. 112, 103350 (2019).
CVAT.ai Corporation. Computer Vision Annotation Tool (CVAT) (2022).
Mullen Jr, J. F., Tanner, F. R. & Sallee, P. A. Comparing the effects of annotation type on machine learning detection performance. In: Proceedings of the ieee/cvf conference on computer vision and pattern recognition workshops, 0–0 (2019).
Goodfellow, I., Bengio, Y. & Courville, A. Deep learning (MIT press, 2016).
Oksuz, K., Cam, B. C., Kalkan, S. & Akbas, E. Imbalance problems in object detection: A review. IEEE Trans. Pattern Anal. Mach. Intell. 43, 3388–3415 (2020).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, 2980–2988 (2017).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: Unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 779–788 (2016).
Pan, S. J. & Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2009).
Itseez. Open source computer vision library. https://github.com/itseez/opencv (2015).
Sabottke, C. F. & Spieler, B. M. The effect of image resolution on deep learning in radiography. Radiol. Artif. Intell. 2, e190015 (2020).
Thambawita, V. et al. Impact of image resolution on deep learning performance in endoscopy image classification: an experimental study using a large dataset of endoscopic images. Diagnostics 11, 2183 (2021).
Wood, L. et al. Kerascv. https://github.com/keras-team/keras-cv (2022).
Sarma, U. C., Winship, A. L. & Hutt, K. J. Comparison of methods for quantifying primordial follicles in the mouse ovary. J. Ovarian Res. 13, 1–11 (2020).
Litjens, G. et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286 (2016).
Pham, H. H. N. et al. Detection of lung cancer lymph node metastases from whole-slide histopathologic images using a two-step deep learning approach. Am. J. Pathol. 189, 2428–2439 (2019).
Zadeh Shirazi, A. et al. Deepsurvnet: deep survival convolutional network for brain cancer survival rate classification based on histopathological images. Med. Biol. Eng. Comput. 58, 1031–1045 (2020).
Zhao, K. et al. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. EBioMedicine61 (2020).
Sun, C. et al. Deep learning with whole slide images can improve the prognostic risk stratification with stage iii colorectal cancer. Comput. Methods Programs Biomed. 221, 106914 (2022).
Saillard, C. et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 72, 2000–2013 (2020).
Liu, H. & Kurc, T. Deep learning for survival analysis in breast cancer with whole slide image data. Bioinformatics 38, 3629–3637 (2022).
Zheng, Q. et al. Accurate diagnosis and survival prediction of bladder cancer using deep learning on histological slides. Cancers 14, 5807 (2022).
Rai, A. Explainable ai: From black box to glass box. J. Acad. Mark. Sci. 48, 137–141 (2020).
Yang, G., Ye, Q. & Xia, J. Unbox the black-box for the medical explainable ai via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Information Fusion 77, 29–52 (2022).
Padmanabhan, D. C., Plöger, P. G., Arriaga, O. & Valdenegro-Toro, M. Dext: Detector explanation toolkit. In: World Conference on Explainable Artificial Intelligence, 433–456 (Springer, 2023).
Petsiuk, V. et al. Black-box explanation of object detectors via saliency maps. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 11443–11452 (2021).
Karasmanoglou, A., Antonakakis, M. & Zervakis, M. Heatmap-based explanation of yolov5 object detection with layer-wise relevance propagation. In: 2022 IEEE International Conference on Imaging Systems and Techniques (IST), 1–6 (IEEE, 2022).
Gudovskiy, D., Hodgkinson, A., Yamaguchi, T., Ishii, Y. & Tsukizawa, S. Explain to fix: A framework to interpret and correct dnn object detector predictions. arXiv preprint arXiv:1811.08011 (2018).
Acknowledgements
This research was funded with the support of the Institut National de la Recherche pour I’Agriculture, I’Alimentation et I’Environnement (INRAE), Phase department innovative project, and Bill & Melinda Gates Foundation CONTRABODY grant. We also acknowledge the support received from the dedicated team in the rodent animal facility, experimental unit: UEPAO (PAO, INRAE: Animal Physiology Facility https://doi.org/10.15454/1.5573896321728955E12) for their attention to details and commitment to animal welfare. This work also benefited from the equipment and expertise of the imaging facility Plateau d’Imagerie Cellulaire (PIC) of UMR-PRC.
Author information
Authors and Affiliations
Contributions
M.HH. prepared samples, performed annotations, generated figures, analyzed results, and wrote original manuscript. E.R. obtained funding, supervised, reviewed and edited the manuscript. M.R. obtained funding, conceptualized, developed models, analyzed results, supervised, and reviewed and edited the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hassan, M.H., Reiter, E. & Razzaq, M. Automatic ovarian follicle detection using object detection models. Sci Rep 14, 31856 (2024). https://doi.org/10.1038/s41598-024-82904-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-82904-8