Abstract
The accurate preoperative staging of laryngeal squamous cell carcinoma (LSCC) provides valuable guidance for clinical decision-making. The objective of this study was to establish a multiparametric MRI model using radiomics and deep learning (DL) to preoperatively distinguish between Stages I–II and III–IV of LSCC. Data from 401 histologically confirmed LSCC patients were collected from two centers (training set: 213; internal test set: 91; external test set: 97). Radiomics features were extracted from the MRI images, and seven radiomics models based on single and combined sequences were developed via random forest (RF). A DL model was constructed via ResNet 18, where DL features were extracted from its final fully connected layer. These features were fused with crucial radiomics features to create a combined model. The performance of the models was assessed using the area under the receiver operating characteristic (ROC) curve (AUC) and compared with the radiologist performances. The predictive capability of the combined model for Progression-Free Survival (PFS) was evaluated via Kaplan–Meier survival analysis and the Harrell’s Concordance Index (C-index). In the external test set, the combined model had an AUC of 0.877 (95% CI 0.807–0.946), outperforming the DL model (AUC: 0.811) and the optimal radiomics model (AUC: 0.835). The combined model significantly outperformed both the DL (p = 0.017) and the optimal radiomics models (p = 0.039), and the radiologists (both p < 0.050). Moreover, the combined model demonstrated great prognostic predictive value in patients with LSCC, achieving a C-index of 0.624 for PFS. This combined model enhances preoperative LSCC staging, aiding in making more informed clinical decisions.
Similar content being viewed by others
Introduction
Laryngeal squamous cell carcinoma (LSCC), a prevalent malignancy in the head and neck, has experienced a 12% increase in incidence and a 24% rise in prevalence over the past 30 years1,2. Despite significant strides in diagnostic methods and therapeutic interventions, survival rates remain stagnant3. The treatment strategy for LSCC is guided by preoperative clinical staging according to the tumor–node–metastasis (TNM) system (8th edition) by the American Joint Committee on Cancer (AJCC). Early-stage LSCC (Stages I–II) is typically treated with laser resection, partial laryngectomy, or radiotherapy, whereas advanced-stage LSCC (Stages III–IV) often requires total laryngectomy combined with chemoradiotherapy4. Studies5,6 have demonstrated a significant discrepancy between the clinical and pathological stages of LSCC, with approximately one-third of patients experiencing stage adjustments post-laryngectomy, which may result in either delayed treatment or overtreatment. This inconsistency is owing to the reliance of the clinical TNM system on the anatomical information from endoscopy and imaging, which fails to capture the heterogeneous nature of a tumor. Additionally, accurately assessing thyroid cartilage invasion is crucial for preoperative staging but remains challenging. Conventional CT and MRI have a limited diagnostic accuracy in detecting thyroid cartilage invasion, often leading to overestimations and unnecessary total laryngectomies. Therefore, developing a novel, noninvasive preoperative assessment method that better aligns clinical and pathological staging is essential for optimizing LSCC treatment strategies.
Radiomics facilitates the extraction of complex imaging features from tumor images and the exploration of intratumoral heterogeneity7. It has been increasingly applied to the tumor grading, prognosis, and lymph node metastasis prediction in head and neck tumor patients8,9,10,11. Liu et al.12 developed a radiomics nomogram using enhanced CT images to differentiate between the T2 and T3 stages in 118 patients with laryngeal and hypopharyngeal squamous cell carcinoma (LHSCC), achieving an area under the curve (AUC) of 0.817. However, traditional radiomics primarily focuses on image features, limiting its ability to capture the full complexity of medical images13. Conversely, deep learning (DL) techniques, as data-driven algorithms, have demonstrated a superior capability in clinical practice14. A study15 established a diffusion-weighted imaging (DWI)-based DL model for forecasting local recurrence in patients with LHSCC (AUC = 0.767). This DL method excels in feature representation, owing to its ability to integrate feature extraction and classification16; however, it is limited due to the poor interpretability of the outcome17. Studies18 have shown that combining these two types of features in a complementary manner can improve results. Chen et al.16 developed a combined radiomics-DL model based on contrast-enhanced CT, achieving an AUC of 0.849 in distinguishing LSCC Stages I–II versus III–IV, surpassing that of individual radiomics (0.704) and DL (0.724) models. However, previous related studies have primarily based on CT images and have encountered limitations due to small sample sizes and the absence of external validation10,12,16,19. The use of MRI to combine radiomics and DL features to analyze the preoperative staging of LSCC has not been investigated thus far.
We hypothesize that a combined model integrating radiomics and DL features based on multiparametric MRI can enable a more accurate preoperative distinction for LSCC patients (Stage I–II and Stage III–IV). This study aims to develop and validate the efficacy of radiomics, DL, and combined models for the preoperative staging of LSCC via a two-center dataset, and to compare their performance with that of the radiologists. Furthermore, we also explored the value of the combined model in prognostic prediction.
Materials and methods
Ethics declarations and consent to participate
This study complies with the Declaration of Helsinki. Due to the retrospective nature of the research, the Clinical Research Ethics Committee of the First Affiliated Hospital of Chongqing Medical University and the Ethics Committee of West China Hospital, Sichuan University jointly waived the requirement for informed consent. This retrospective study was jointly approved by the Ethics Committees of the aforementioned institutions and was conducted in strict accordance with relevant ethical guidelines (approval number: 2020080).
Patient cohort
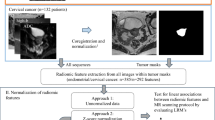
We retrospectively collected MRI images from 401 patients with histologically confirmed LSCC at two centers: the First Affiliated Hospital of Chongqing Medical University (Center 1, n = 304) and West China Hospital of Sichuan University (Center 2, n = 97). The inclusion criteria were as follows: (1) no preoperative treatment; (2) standard multiparametric MRI scans performed within 14 days prior to surgery; and (3) availability of comprehensive clinical data. The exclusion criteria were as follows: (1) tumor recurrence; (2) other concurrent tumors; (3) tumors with lesions too small (the largest cross-sectional diameter was less than 5 mm); and (4) poor image quality that precluded the determination of lesion extent or the presence of severe artifacts. Finally, patients from Center 1 were randomly assigned to a training set (n = 213) and an internal test set (n = 91) in a 7:3 ratio, whereas patients from Center 2 constituted the external test set (n = 97). The patient recruitment process is illustrated in Fig. 1.
The clinical parameters included age, gender, history of drinking and smoking, tumor site, clinical and pathological TNM stage. The tumors were staged according to the guidelines of the AJCC TNM Staging System (8th Edition). Furthermore, according to the pathological TNM stage, the patients were classified into stage I–II patients and stage III–IV patients.
MRI image acquisition
All the patients underwent scans with comparable settings but on different MRI machines (Supplementary Table S1). The scanning range spanned from the base of the skull to the thoracic inlet, employing acquisition sequences that included axial T1-weighted (T1WI) images, fat-suppressed T2-weighted (T2WI) images, and contrast-enhanced T1-weighted fat-suppressed (CE-T1WI-FS) scans.
Traditional radiomics model construction and validation
All original MRI images were anonymized to protect patient confidentiality. Radiologist A (6 years of experience in head and neck diagnosis) used ITK-SNAP 3.6.0 software (www.itk-snap.org) to manually outline the region of interest (ROI) at the tumor margins, layer-by-layer, forming a volume of interest (VOI). Multiplanar reconstruction was used during VOI segmentation to ascertain the extent of the lesion. To ensure the feature reproducibility and stability, radiologist A re-delineated the VOIs for 40 randomly selected patients after a 30-day interval. Additionally, radiologist B (4 years of experience) independently delineated these same cases. The intraclass correlation coefficient (ICC) was computed to assess both intra- and inter-observer agreement, features exhibiting an ICC greater than 0.75 were considered stable and reliable. A senior radiologist (Radiologist C; possessing 24 years of experience) carefully reviewed each VOI, while all radiologists were kept unaware of the clinical information during both the delineation and review processes.
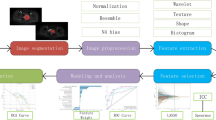
All images were uniformly resampled to a resolution of 1 × 1 × 1 mm and preprocessed via N4 bias field correction and normalization. For feature extraction, PyRadiomics (version 3.0.1) was employed, resulting in the extraction of a comprehensive set of 863 features per sequence. These features encompassed shape, first-order, texture matrices, and advanced features from wavelet transforms and Gaussian‒Laplacian filtering. To minimize overfitting and identify informative features, a methodical selection procedure eliminated features with ICCs lower than 0.75. From pairs of highly correlated features (r > 0.9, based on Spearman’s coefficient), one feature was preserved. Following z-score normalization, the least absolute shrinkage and selection operator (LASSO) regression was employed for dimensionality reduction. Finally, the optimal feature subset was selected via recursive feature elimination (RFE) for model construction.
We developed seven radiomics models via the random forest (RF) algorithm, a powerful and robust ensemble learning technique20,21. The single-sequence models included Radiomics-T1WI, Radiomics-T2WI, and Radiomics-CE-T1WI. The multisequence combination models included Radiomics-CE-T1WI + T1WI + T2WI (referred to as Radiomics-ALL), Radiomics-CE-T1WI + T1WI, Radiomics-CE-T1WI + T2WI, and Radiomics-T1WI + T2WI. To enhance the models’ predictive performance, a grid search algorithm incorporating fivefold cross-validation was employed. Figure 2 illustrates the overall study framework.
The study’s framework is structured as follows: (1) Multi-sequence MRI tumor segmentation; (2) Feature extraction and model Construction, where radiomics features were extracted from three-dimensional segmentation images to develop radiomics models. Deep learning networks were constructed using cropped two-dimensional regions of interest (ROIs), from which DL features were extracted. The crucial radiomics features were then fused with DL features to create a combined model; (3) Radiologist evaluation; and (4) Model performance assessment.
Deep learning model construction and visualization
We selected ResNet-18, a renowned convolutional neural network (CNN) pretrained on ImageNet, for DL model construction owing to its powerful feature extraction ability and ability to retain low-scale features22. From 3D segmentation masks of three MRI sequences, we cropped 2D ROIs encompassing the largest tumor cross-sections as input images. The network generated a probability value at the output, serving as the basis for decision-making. Prior to input, the tumor images were resized to a uniform dimension of 224 × 224 pixels and normalized using the z-score method. To mitigate overfitting, we applied online data augmentation methods, including random horizontal and vertical flips, and random cropping. During training, the stochastic gradient descent (SGD) optimizer was utilized to adjust the model parameters. Additionally, we employed gradient-weighted class activation mapping (Grad-CAM) to provide a visualization of the model, by integrating gradient data obtained from the final layer of ResNet-18 to generate class activation maps. The training process was performed on NVIDIA GeForce RTX 3090 GPU, with network frameworks built via PyTorch and CUDA for accelerated computation.
Construction of the combined model and radiologists’ visual evaluations
A combined model was constructed that integrates radiomics and DL features through these steps: First, radiomics features with an ICC value exceeding 0.75 were identified as crucial features during the construction of the radiomics model utilizing CE-T1WI + T1WI + T2WI. Second, DL features were extracted from the final fully connected layer, which serves as a feature extractor at the end of the network. Finally, the crucial radiomics features were integrated with the extracted DL features to form a combined dataset. After applying LASSO and RFE for feature selection, the optimal number of features corresponding to the minimum normalized mean squared error (NMSE) was selected via fivefold cross-validation, and a combined model was subsequently constructed using RF.
To evaluate the efficacy of machine learning models in comparison to radiologists’ visual assessments during the preoperative staging of LSCC, we recruited two radiologists: Doctor 1 (6 years of experience) and Doctor 2 (5 years of experience), neither of them participated in the segmentation procedure. Each radiologist assessed the multi-sequence MRI images (coronal and sagittal views) of patients in the external test set without prior pathological information. Based on the AJCC TNM staging system (8th edition), they classified patients into two preoperative staging categories: Stages I–II and III–IV. This categorization was based on assessments of tumor morphology, size, margins, thyroid cartilage invasion23 (suspected when the signal intensity or degree of enhancement of the cartilage resembles that of the adjacent tumor), and cervical lymph node metastasis16 (jugulodigastric nodes measuring ≥ 1.5 cm, or lymph nodes outside the jugulodigastric chain ≥ 1.0 cm, coupled with irregular margins and central parenchymal necrosis, suggest a high probability of metastasis).
Statistical analysis
Continuous variables were analyzed via the Student’s t-test or Mann–Whitney U test. Categorical variables were analyzed with the chi-square test or Fisher’s exact test. The interobserver agreement between Doctor 1 and Doctor 2 was evaluated using Cohen’s Kappa test. To quantify model performance, the following metrics were calculated: the AUC, F1-score, Accuracy, Sensitivity (Recall), Specificity, Positive Predictive Value (PPV), and Negative Predictive Value (NPV). Significant differences in AUC among the models were assessed using the DeLong test, with a p-value < 0.05 considered statistically significant. Calibration curves were employed to evaluate the consistency between the predictions of the optimal model and actual observations. The combined model’s prognostic ability for predicting progression-free survival (PFS) was analyzed using Harrell’s concordance index (C-index) and Kaplan–Meier methods. All the analyses were conducted via SPSS (v26.0) and Python (v3.8).
Results
Clinical characteristics
Table 1 presents the baseline clinical characteristics of all patients. This study included 401 participants, comprising 384 males (95.8%) and 17 females (4.2%). The statistical analysis indicated that there were no significant differences across the three patient groups regarding the pathological TNM stage, clinical TNM stage (including T stage and N stage), gender, age, smoking status, drinking status, tumor site, or differentiation (all p > 0.05). The clinical characteristics were consistently distributed across the three cohorts, ensuring the reliability of the model prediction results.
Feature subset selection and model construction
For the radiomics model, 863 features were extracted from each sequence image. After excluding features with an ICC of less than 0.75, a Spearman correlation analysis was conducted on the features that remained, resulting in the retention of 672 features (including 230 from T1WI, 241 from T2WI, and 201 from CE-T1WI). Subsequent feature selection via the LASSO and RFE algorithms led to the identification of the top 15 radiomics features from both individual sequences and the combined sequence for the construction of the radiomics models. ResNet18 was employed to construct the DL model. Additionally, 2048 DL features were extracted from the network’s final fully connected layer. These features were then fused with the crucial radiomics features to select the optimal hybrid feature set, which included 10 radiomics features and 5 DL features. The SHapley Additive exPlanations (SHAP) methodology was employed to visualize the impact of these features on the combined model (Fig. 3). DL214, located at the top of the SHAP summary plot, indicating its dominant impact on model predictions.
The SHAP plot of the top 15 features in the combined model, with feature value indicating the importance of predictive features. SHAP value serve as a unified measure of the impact of a particular feature in the model. Red dots represent positive correlations, while blue dots represent negative correlations. DL, deep learning; glcm, gray-level co-occurrence matrix; gldm, gray level dependence matrix; glszm, gray-level size zone matrix.
Performance assessment of different models
Supplementary Table S2 and Table 2 present the predictive performance of each model. Out of the seven radiomics models analyzed, the Radiomics-ALL model demonstrated superior performance in both test sets, with AUCs of 0.857 (95% CI 0.778–0.936) and 0.835 (95% CI 0.756–0.915), respectively. This model was selected as the optimal radiomics model for subsequent comparisons with both the DL and the combined model.
In both test sets, the combined model demonstrated significantly greater predictive performance than both the Radiomics-ALL model and the DL model, with AUCs of 0.913 (95% CI 0.852–0.974) and 0.877 (95% CI 0.807–0.946), respectively. The findings from the DeLong test conducted in the external test set (Supplementary Table S3) revealed that the combined model significantly surpassed both the Radiomics-ALL model (p = 0.039) and the DL model (p = 0.017). Additionally, the Radiomics-ALL model demonstrated enhanced performance when compared to the DL model (p = 0.040). In the external test set, the combined model, Radiomics-ALL model, and DL model achieved accuracies of 0.814, 0.814, and 0.732, respectively. Their specificities were 0.711, 0.689, and 0.733, while sensitivities (recall) reached 0.904, 0.923, and 0.730, respectively. The F1-scores for these models were 0.839, 0.842, and 0.745, respectively. The analysis of the calibration curves demonstrated that the combined model’s predictive capabilities align closely with the ideal model’s performance in both test sets. (Fig. 4b). Furthermore, Grad-CAM Heatmaps (Fig. 5) presents the class activation maps, which visually highlight the significant regions in Stages I–II and III–IV of LSCC. The red areas emphasize the regions most critical to ResNet-18’s classification process, primarily located at the tumor boundaries and within intratumoral regions.
Performance comparison of three models and radiologists in the external test set. (a) Receiver Operating Characteristic (ROC) curves for the combined model, the Radiomics-ALL model, the Deep Learning (DL) model, and two radiologists. Numbers in parentheses indicate the respective areas under the ROC curves. (b) Calibration curve analysis for the combined model. The solid line demonstrates the performance of the combined model; the closer it is to the dashed diagonal line, the higher the consistency between predicted probabilities and observed probabilities.
Performance comparison of the three models with radiologists
We compared the performance of the aforementioned models in the external test with the visual assessments of two radiologists. The diagnostic outcomes from both radiologists demonstrated a good agreement (kappa coefficient > 0.7); the AUCs were 0.716 (95% CI 0.614–0.816) and 0.717 (95% CI 0.616–0.818), respectively. The combined model and the Radiomics-ALL model significantly outperformed both radiologists (p = 0.016 and p < 0.001 for Doctor 1; p = 0.017 and p < 0.001 for Doctor 2). However, no significant difference was observed in the diagnostic efficacy between the DL model and the two radiologists (p = 0.078 for Doctor 1; p = 0.075 for Doctor 2). The ROC curve for the aforementioned models is shown in Fig. 4a. The confusion matrices (Fig. 6) reveal that the two radiologists misclassified 18/52 and 19/52 pathologically confirmed Stage III–IV cases as Stage I–II, respectively. In contrast, the combined model reduced misclassification to 5/52 advanced-stage cases, significantly lowering undertreatment risk (p < 0.05, McNemar’s test).
Follow-up study and survival analysis
A cohort of 267 patients was followed until November 30, 2023, with progression-free survival (PFS) defined as the duration from the date of surgery to the occurrence of either demise, metastasis, or recurrence24. Patients without any events were censored. The median PFS was 38 months, ranging from 3 to 71 months. Using the Youden index-derived optimal cutoff, patients were stratified into low-risk and high-risk groups. The combined model’s prognostic utility was assessed by correlating predicted risk scores with actual clinical outcomes. The model demonstrated moderate discriminative ability for PFS (C-index = 0.624, 95% CI 0.526–0.722). Kaplan–Meier analysis revealed significantly prolonged PFS in low-risk versus high-risk patients (hazard ratio [HR] = 0.454, 95% CI 0.241–0.856, p = 0.012) (Fig. 7).
Discussion
This study developed multiple radiomics models, a DL model, and a combined model using multiparametric MRI to improve the accuracy of preoperative staging for patients with LSCC (Stage I–II versus Stage III–IV). We also compared the diagnostic performances of the two radiologists with those of the models. The combined model demonstrated the best classification performance in both the internal (AUC = 0.913, 95% CI 0.852–0.974) and external test sets (AUC = 0.877, 95% CI 0.807–0.946), outperforming the other models and radiologists, thus highlighting its potential as a reliable predictor for guiding clinical decision-making.
The surgical strategies differ for early-stage LSCC (Stages I–II) and advanced-stage LSCC (Stages III–IV). Patients who undergo total laryngectomy will face greater suffering and increased burdens on their daily lives, whereas partial laryngectomy can preserve patients’ vocal cords and enhance their quality of life. Therefore, precise preoperative staging plays a vital role in determining suitable treatment strategies for individuals diagnosed with LSCC. The preoperative TNM staging of LSCC largely relies on imaging examination results. However, the diagnostic accuracy of radiologists heavily depends on their experience. In this study, the AUCs of the two radiologists in the external test set were lower than those of the combined, Radiomics-ALL, and DL models, indicating insufficient accuracy in their preoperative evaluations for LSCC patients—a finding consistent with prior research25. For example, Wang et al.19 reported radiologists achieving an AUC of 0.775 in distinguishing T3/T4 stages via CT, which underperformed both radiomics (AUC = 0.862) and a combined nomogram (AUC = 0.892), likely due to limitations of visual assessment. Further analysis of 97 patients in the external test revealed that radiologists’ underdiagnosis of advanced LSCC (stages III–IV) contributed to undertreatment, such as incomplete surgeries, whereas the combined model demonstrated superior staging accuracy. These results underscore the potential of the combined model to enhance personalized clinical decision-making by improving diagnostic precision and guiding appropriate therapeutic interventions.
The application of radiomics and DL in medical image diagnosis has introduced innovative and advanced methodologies for clinical practice26,27. Radiomic features offer insights into the tumor microenvironment, complementing clinical data. However, previous radiomics studies of LSCC have primarily relied on CT images. Conversely, this study leveraged the discriminatory advantages of MRI in identifying abnormalities in soft tissue and cartilage and constructed multiple radiomics models with different combinations26. Studies17,28 have shown that the combination of various MRI sequences offers a more comprehensive understanding of tumor characteristics. Ding et al.29 found that a radiomics model incorporating T2WI, DCE-T1WI, and apparent diffusion coefficient (ADC) maps was more effective in predicting Ki-67 expression in early endometrial cancer preoperatively, achieving an AUC of 0.852 on the validation set, compared to single-sequence models. In this study, the Radiomics-ALL model based on CE-T1WI + T1WI + T2WI achieved the best performance among the seven radiomics models; this confirmed that multiple sequences can provide more information regarding the tumor area and improve the ability to differentiate, especially as T2WI can better delineate the lesion contour, whereas CE-T1WI can indicate the blood supply of the tumor17,30. Extracting more comprehensive features from combined sequences helps improve the diagnostic efficacy of the model31. Additionally, we developed a DL prediction model that identifies key features directly from raw pixel data of input images to perform classification and prediction tasks in an end-to-end manner32.While prior studies suggest DL typically outperforms radiomics in classification tasks33, our results showed the DL model underperformed in comparison to the Radiomics-ALL model (p = 0.040). This discrepancy may stem from initializing the DL model with weights pretrained on ImageNet rather than tumor-specific imagery34, coupled with DL’s inherent reliance on large datasets—a limitation that can compromise performance with smaller sample sizes18. Furthermore, the DL model exhibited limited generalizability, as its diagnostic ability did not significantly differ from that of the two radiologists, highlighting challenges in translating theoretical DL advantages into practical clinical utility without domain-specific optimization and sufficient training data.
Notably, integrating radiomics with DL presents promising opportunities for clinical research. For instance, Lan et al.35 developed 17 predictive models for occult cervical lymph node metastasis (OCLNM) in early-stage oral cancer and LCSS, among which a ResNet50 model combining radiomics and DL achieved superior performance, demonstrating an AUC of 0.834 in external validation Set 2. This underscores the complementary strengths of DL and radiomics. Building on this concept, we developed a combined model to improve preoperative staging for LSCC patients. Our combined approach achieved the highest predictive accuracy in preoperative LSCC staging, outperforming both conventional radiomics and standalone DL models.
Among the optimal features, 10 were radiomics-based (8 wavelet transform features and 2 original features), and 5 were DL-derived. The prominence of wavelet-transform features likely stems from their ability to capture coarse-grained textural information, enhancing tumor heterogeneity representation in images33,36 and aiding LSCC staging12. Radiomics features like gray-level co-occurrence matrix (GLCM), which identifies discrete changes in structural characteristics of tissues, and gray-level size zone matrix (GLSZM), which quantifies homogeneous voxel clusters, correlate with tumor heterogeneity37. These features provide clinically interpretable insights into tumor biology, such as invasion patterns, necrosis, and neovascularity16,38,39, aligning with LSCC staging criteria10,12. SHAP analysis identified DL214 as the most impactful feature, indicating its extraction of complex patterns—like tumor margins and intratumoral heterogeneity—through automated DL processing32. Grad-CAM visualizations highlighted tumor boundaries and internal structures as critical decision-making regions, suggesting that DL features (e.g., DL214) captured morphological characteristics distinguishing early-stage (I–II) from advanced-stage (III–IV) LSCC16. Advanced tumors may exhibit more irregular shapes, blurred margins, and greater tumor heterogeneity, mirroring radiologists’ diagnostic criteria5,40 (Fig. 5). This clinical alignment validates the model’s relevance. In summary, the integration of DL and radiomics synergistically enhances predictive modeling: DL extracts intricate imaging patterns, while radiomics provides their quantitative analysis. This combined approach captures diverse data dimensions, improving predictive accuracy. The model’s superior performance in LSCC preoperative staging highlights the effectiveness of merging DL and radiomics for precise, robust predictions. Furthermore, its ability to stratify patients into high- and low-risk PFS categories underscores its clinical utility as a prognostic tool.
We must acknowledge the limitations of our study. First, this research was limited by its retrospective nature; a prospective study would provide stronger evidence for the predictive ability of our model. Second, multicenter MRI data, acquired from scanners with varying parameters, introduced inter-scanner variability affecting radiomic feature consistency despite efforts to standardize image quality. Advanced harmonization techniques are critical to address this challenge, with ComBat demonstrating effectiveness in reducing feature variability across centers and CycleGAN offering complementary domain adaptation to harmonize images from heterogeneous devices41,42. Additionally, incorporating functional MRI sequences like DWI and ADC could provide novel insights into tumor staging by quantitating cellular-level water diffusion, addressing conventional MRI’s limitations. However, these sequences were unavailable for some patients and thus excluded from the current analysis. Future studies will systematically integrate these functional metrics to enhance staging precision. Finally, DL models require extensive data for training. Thus, incorporating images from more patients to improve the model’s ability to generalize is within the scope of our ongoing research.
Conclusion
This study demonstrates that a combined model, which integrates radiomics and DL features based on multiparametric MRI, exhibits a superior predictive capability in differentiating between Stages I–II and III–IV of LSCC. This development has the potential to improve clinical decision-making, thereby leading to better patient outcomes.
Data availability
Upon receipt of a reasonable request, the datasets employed and/or examined during the present study can be obtained from the corresponding author.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 74, 229–263. https://doi.org/10.3322/caac.21834 (2024).
Nocini, R., Molteni, G., Mattiuzzi, C. & Lippi, G. Updates on larynx cancer epidemiology. Chin. J. Cancer Res. 32, 18–25. https://doi.org/10.21147/j.issn.1000-9604.2020.01.03 (2020).
Chen, L., Wang, H., Zeng, H., Zhang, Y. & Ma, X. Evaluation of CT-based radiomics signature and nomogram as prognostic markers in patients with laryngeal squamous cell carcinoma. Cancer Imaging https://doi.org/10.1186/s40644-020-00310-5 (2020).
Obid, R., Redlich, M. & Tomeh, C. The treatment of laryngeal cancer. Oral Maxillofac. Surg. Clin. North Am. 31, 1–11. https://doi.org/10.1016/j.coms.2018.09.001 (2019).
Itamura, K., Hsue, V. B., Barbu, A. M. & Chen, M. M. Diagnostic assessment (Imaging) and staging of laryngeal Cancer. Otolaryngol. Clin. North Am. 56, 215–231. https://doi.org/10.1016/j.otc.2022.12.006 (2023).
Contrera, K. J. et al. Clinical versus pathologic laryngeal cancer staging and the impact of stage change on outcomes. Laryngoscope 131, 559–565. https://doi.org/10.1002/lary.28924 (2020).
Avanzo, M. et al. Machine and deep learning methods for radiomics. Med. Phys. https://doi.org/10.1002/mp.13678 (2020).
Preda, L. et al. Diagnostic accuracy of surface coil MRI in assessing cartilaginous invasion in laryngeal tumours: Do we need contrast-agent administration?. Eur. Radiol. 27, 4690–4698. https://doi.org/10.1007/s00330-017-4840-x (2017).
Guo, R. et al. CT-based radiomics features in the prediction of thyroid cartilage invasion from laryngeal and hypopharyngeal squamous cell carcinoma. Cancer Imaging https://doi.org/10.1186/s40644-020-00359-2 (2020).
Ren, J. et al. Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I-II and III-IV head and neck squamous cell carcinoma. Eur. J. Radiol. 106, 1–6. https://doi.org/10.1016/j.ejrad.2018.07.002 (2018).
Zheng, Y.-M. et al. A CT-based deep learning radiomics nomogram to predict histological grades of head and neck squamous cell carcinoma. Acad. Radiol. 30, 1591–1599. https://doi.org/10.1016/j.acra.2022.11.007 (2023).
Liu, Q. et al. Machine learning model to preoperatively predict T2/T3 staging of laryngeal and hypopharyngeal cancer based on the CT radiomic signature. Eur. Radiol. 34, 5349–5359. https://doi.org/10.1007/s00330-023-10557-8 (2024).
Swati, Z. N. K. et al. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 75, 34–46. https://doi.org/10.1016/j.compmedimag.2019.05.001 (2019).
Xu, Z.-H. et al. Computer-aided diagnosis of laryngeal cancer based on deep learning with laryngoscopic images. Diagnostics https://doi.org/10.3390/diagnostics13243669 (2023).
Tomita, H. et al. Deep learning approach of diffusion-weighted imaging as an outcome predictor in laryngeal and hypopharyngeal cancer patients with radiotherapy-related curative treatment: A preliminary study. Eur. Radiol. 32, 5353–5361. https://doi.org/10.1007/s00330-022-08630-9 (2022).
Chen, X. et al. A combined model integrating radiomics and deep learning based on contrast-enhanced CT for preoperative staging of laryngeal carcinoma. Acad. Radiol. 30, 3022–3031. https://doi.org/10.1016/j.acra.2023.06.029 (2023).
Hagiwara, A. et al. From simultaneous rapid acquisition methods and analysis techniques using scoring, machine learning, radiomics, and deep learning to the generation of novel metrics. Invest. Radiol. https://doi.org/10.1097/rli.0000000000000962 (2023).
Wu, X. et al. Exploring the predictive value of additional peritumoral regions based on deep learning and radiomics: A multicenter study. Med. Phys. 48, 2374–2385. https://doi.org/10.1002/mp.14767 (2021).
Wang, F. et al. Radiomic nomogram improves preoperative t category accuracy in locally advanced laryngeal carcinoma. Front. Oncol. https://doi.org/10.3389/fonc.2019.01064 (2019).
Zhou, J. et al. Diagnosis of benign and malignant breast lesions on DCE-MRI by using radiomics and deep learning with consideration of peritumor tissue. J. Magn. Reson. Imaging 51, 798–809. https://doi.org/10.1002/jmri.26981 (2019).
Lefebvre, T. L. et al. Development and validation of multiparametric MRI–based radiomics models for preoperative risk stratification of endometrial cancer. Radiology 305, 375–386. https://doi.org/10.1148/radiol.212873 (2022).
Liang, G. & Zheng, L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Progr. Biomed. https://doi.org/10.1016/j.cmpb.2019.06.023 (2020).
Park, C. J. et al. Preoperative MRI evaluation of thyroid cartilage invasion in patients with laryngohypopharyngeal cancer: Comparison of contrast-enhanced 2D spin-echo and 3D T1-weighted radial gradient recalled-echo techniques. Am. J. Neuroradiol. 42, 1690–1694. https://doi.org/10.3174/ajnr.A7213 (2021).
Rizzo, M. et al. Radiotherapy plus pembrolizumab for advanced urothelial carcinoma: Results from the ARON-2 real-world study. Sci. Rep. https://doi.org/10.1038/s41598-024-70182-3 (2024).
Gitto, S. et al. MRI radiomics-based machine learning for classification of deep-seated lipoma and atypical lipomatous tumor of the extremities. Radiol. Med. (Torino) 128, 989–998. https://doi.org/10.1007/s11547-023-01657-y (2023).
Allegra, E. et al. Early glottic cancer: Role of MRI in the preoperative staging. Biomed. Res. Int. 1–7, 2014. https://doi.org/10.1155/2014/890385 (2014).
Lambin, P. et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762. https://doi.org/10.1038/nrclinonc.2017.141 (2017).
Chen, W. et al. Prediction of the Ki-67 expression level in head and neck squamous cell carcinoma with machine learning-based multiparametric MRI radiomics: A multicenter study. BMC Cancer https://doi.org/10.1186/s12885-024-12026-x (2024).
Ding, S.-X. et al. Radiomics model based on multi-sequence MRI for preoperative prediction of ki-67 expression levels in early endometrial cancer. Sci. Rep. https://doi.org/10.1038/s41598-023-49540-0 (2023).
Li, J. et al. Comparison ofMRIandCT-based radiomics and their combination for early identification of pathological response to neoadjuvant chemotherapy in locally advanced gastric cancer. J. Magn. Reson. Imaging 58, 907–923. https://doi.org/10.1002/jmri.28570 (2022).
Li, Q. et al. The effect of magnetic resonance imaging based radiomics models in discriminating stage I-II and III–IVa nasopharyngeal carcinoma. Diagnostics https://doi.org/10.3390/diagnostics13020300 (2023).
Yu, Q. et al. Deep learning-assisted diagnosis of benign and malignant parotid tumors based on contrast-enhanced CT: A multicenter study. Eur. Radiol. 33, 6054–6065. https://doi.org/10.1007/s00330-023-09568-2 (2023).
Papadimitroulas, P. et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Med. 83, 108–121. https://doi.org/10.1016/j.ejmp.2021.03.009 (2021).
Chen, W. et al. CT-based deep learning radiomics signature for the preoperative prediction of the muscle-invasive status of bladder cancer. Front. Oncol. https://doi.org/10.3389/fonc.2022.1019749 (2022).
Lan, T. et al. MRI-based deep learning and radiomics for prediction of occult cervical lymph node metastasis and prognosis in early-stage oral and oropharyngeal squamous cell carcinoma: A diagnostic study. Int. J. Surg. https://doi.org/10.1097/js9.0000000000001578 (2024).
Rizzo, S. et al. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. https://doi.org/10.1186/s41747-018-0068-z (2018).
Sun, K. et al. Multi-phase contrast-enhanced magnetic resonance image-based radiomics-combined machine learning reveals microscopic ultra-early hepatocellular carcinoma lesions. Eur. J. Nucl. Med. Mol. Imaging 49, 2917–2928. https://doi.org/10.1007/s00259-022-05742-8 (2022).
van Helden, E. J. et al. Radiomics analysis of pre-treatment [(18)F]FDG PET/CT for patients with metastatic colorectal cancer undergoing palliative systemic treatment. Eur. J. Nucl. Med. Mol. Imaging 45, 2307–2317. https://doi.org/10.1007/s00259-018-4100-6 (2018).
Fahmy, A. S. et al. Radiomics and deep learning for myocardial scar screening in hypertrophic cardiomyopathy. J. Cardiovasc. Magn. Reson. 24, 40. https://doi.org/10.1186/s12968-022-00869-x (2022).
Talaat, F. M., Gamel, S. A., El-Balka, R. M., Shehata, M. & ZainEldin, H. Grad-CAM enabled breast cancer classification with a 3D inception-ResNet V2: Empowering radiologists with explainable insights. Cancers https://doi.org/10.3390/cancers16213668 (2024).
Orlhac, F. et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur. Radiol. 31, 2272–2280. https://doi.org/10.1007/s00330-020-07284-9 (2021).
Uema, R. et al. A novel artificial intelligence-based endoscopic ultrasonography diagnostic system for diagnosing the invasion depth of early gastric cancer. J. Gastroenterol. 59, 543–555. https://doi.org/10.1007/s00535-024-02102-1 (2024).
Acknowledgements
We extend our heartfelt gratitude to all the participants in this research.
Funding
This work was supported by the Foundation of Science and Technology Bureau of Yuzhong District, Chongqing, China (Grant No. 20190111), the Natural Science Foundation of Chongqing, China (Grant No. cstc2021jcyj-msxmX0020).
Author information
Authors and Affiliations
Contributions
K Xie and H Jiang contributed equally to this study. Conceived and designed the analysis: K Xie, H Jiang, X Chen, J Peng, L Xu; Provision of study materials or patients: J Peng, F Lv, Q Yue; Collected the data: K Xie, H Jiang, Y Zhou, R Liu; Contributed data or analysis tools: L Xu, K Xie, H Jiang, X Chen, Y Ning, Q Yu, J Peng. Writing—review and editing: K Xie, H Jiang, X Chen, Y Ning, J Peng; All Authors Approve the Final Version of the Manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, K., Jiang, H., Chen, X. et al. Multiparameter MRI-based model integrating radiomics and deep learning for preoperative staging of laryngeal squamous cell carcinoma. Sci Rep 15, 16239 (2025). https://doi.org/10.1038/s41598-025-01270-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-01270-1