Abstract
Hypoxia causes the occurrence of right heart hypertrophy and right heart failure. However, the yak living in the hypoxic environment, does not exhibit hypoxia-related pathological features. Therefore, It is of great significance to explore the hypoxia adaptation mechanism of yak heart. In this study, the yak heart coronary vascular smooth muscle cells (CASMCs) were treated with 21% O2 (normoxic group) and 5% O2 (hypoxic group). The results showed that hypoxia could promote the proliferation of CASMCs. Subsequently, we sequenced CASMCs in normoxic and hypoxic groups. The analysis revealed differential expression of 835 mRNAs, 285 lncRNAs and 126 miRNAs were between the two groups. GO and KEGG analysis showed that the differentially expressed genes were predominantly associated with extracellular matrix components, transcription factor activity, protein binding, immune system processes, metabolic processes and cell development processes and TGF-β, MAPK, cAMP, mTOR, PI3K-Akt and other signaling pathways. By constructing a network of mRNAs, miRNAs and lncRNAs based on the major differentially expressed RNAs, core regulatory elements associated with hypoxic adaptive function were identified. Our study may help to prove the potential role of differential genes related to hypoxic adaptation, and enhanced understanding of the molecular mechanisms of hypoxic adaptation in yak heart.
Similar content being viewed by others
Introduction
In China, the Qinghai-Tibet Plateau, boasting an average altitude exceeding 4000 m, is characterized by severe and complex plateau climate conditions, including low atmospheric pressure, low oxygen content and low temperature1. The heart serves as the driving force of blood circulation, with the coronary arteries acting as vital vessels for heart nutrition. Consequently, the coronary arteries are particularly susceptible to hemodynamic alterations compared to arteries in other organs. Numerous studies have shown that the migration of typical lowland animals to plateau environments often leads to symptoms such as pulmonary hypertension and right heart hypertrophy2,3. Yaks, as a representative animal living in high altitude regions for a long time, despite long-term exposure to such extreme environments. They exhibit remarkable resilience, avoiding clinical symptoms such as right heart hypertrophy and right heart failure through long-term anatomical and physiological adaptations in their cardiac system4,5. Therefore, research on the molecular mechanism of its adaptation to hypoxia has emerged as a long-term discussion hotspot6,7.
Hypoxia has a complex association with several major human diseases, including cardiovascular and cerebrovascular diseases, sleep apnea hypopnea syndrome and altitude sickness8,9. Research have shown that yaks and Tibetans have been found to maintain normal hemoglobin levels, distinguishing them from lowland cattle and lowland humans, suggesting the presence of different molecular mechanisms that protect them from high altitude polycythemia10,11. Consequently, unraveling the molecular genetic foundations of adaptability stands as a central objective in evolutionary genetics. By understanding how plateau animals cope with challenges such as hypoxia and cold, we have gained valuable insights into the evolutionary process of adaptability. Recent strides in transcriptome sequencing technology have opened up new avenues for exploring the molecular genetic basis of adaptive physiological traits.
A growing body of evidence suggests that new mechanisms by which non-coding RNAs interact with mRNAs, including long non-coding RNAs (lncRNAs), messenger RNAs (mRNAs) and microRNAs (miRNAs)12,13,14. LncRNAs, comprising hundreds to thousands of nucleotides, emerge as pivotal players in various biological processes, spanning cell cycle regulation, epigenetic modifications, and energy homeostasis15. Research has shown that lncRNAs act as competitive endogenous RNAs (ceRNAs) and miRNA sponges to regluate the expression of target genes16. Furthermore, lncRNA ENST00113 has been implicated in promoting the growth and metastasis of atherosclerotic cells through the PI3K/Akt/mTOR pathway17. miRNAs consist of small RNA molecules approximately 22 nucleotides in length. Research has shown that miRNAs are involved in the hypoxic remodeling of coronary artery smooth muscle cells (CASMCs) by regulating the transcription levels of transcription factors and genes18. Previous studies have performed comparative analyses of tissue differences between yaks and yellow cattle using genome and transcriptome sequencing techniques. Notably, lung and heart emerged as two pivotal organs showing significant differences, with more than 90% of differentially expressed genes across altitudes demonstrating nonlinear regulation in lung tissue. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed abundant pathways related to cell survival and proliferation, including PI3K-Akt, HIF-1α and ECM-receptor interactions19,20. Those results underscored adaptive transcriptional changes in yak heart and lung tissues.
Therefore, to investigate the regulatory changes of genes associated with hypoxic adaptation in yak heart vessels in depth, we conducted microarray analysis on CASMCs of yak heart under hypoxic (5% O2) and normoxic (21% O2) conditions. Gene Ontology (GO) and KEGG analysis, we identified differentially expressed genes and enriched signaling pathways. Subsequently, we constructed differential gene expression networks for mRNAs, miRNAs and lncRNAs to pinpoint RNA markers linked to hypoxic adaptation in yak heart vessels. This study not only advances our understanding of the molecular mechanisms of hypoxic adaptation in yaks but also facilitates the scientific exploitation of yak material resources.
Results
Culture and identification of yak heart CASMCs
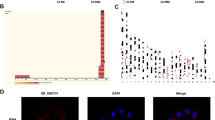
Primary CASMCs were isolated and cultured in vitro. As shown in Fig. 1A, the cultured cells began to adhere to the extension at the bottom of the culture flask within 5 days of the initial culture, and after confluence (7–8 days of culturing) they exhibited a typical “hills and valleys” appearance. The purification of primary cells was confirmed through strongly positive staining for α-smooth muscle actin (green) and calponin-1 (green), but negative staining for CD31 (Fig. 1B). Therefore, IF staining confirmed that more than 98% of the isolated cells were indeed smooth muscle cells.
Effects of hypoxia on the proliferation of CASMCs
The CCK-8 cell proliferation kit was used to measure cell proliferation showed that compared with the normoxic group, the rate of the hypoxic group was significantly higher, and the difference was significant after 48 h (Fig. 2A). In addition, the expression of Proliferating Cell Nuclear Antigen (PCNA) in CASMCs under hypoxic and normoxic conditions was examined by western-blot. The results showed that compared with normoxic condition, hypoxic promoted the expression of PCNA protein (Fig. 2B, C).
Quality detection of total RNA in CASMCs
The RNA extracted from cells of both the normoxic and hypoxic groups underwent agarose electrophoresis, with the results depicted in Fig. 3A. All RNA samples exhibited three distinct bands, 28 S, 18 S and 5 S from top to bottom. Respectively, signifying the absence of notable RNA degradation. This observation indicates that all samples successfully passed quality inspection, meeting the criteria for database construction.
Whole transcriptome sequencing results
This study involved the successful construction of six cDNA libraries using Illumina sequencing technology. Three libraries were derived from the hypoxic group (5% O2 CASMCs), labeled as 5% O2 CASMC-1, 5% O2 CASMC-2 and 5% O2 CASMC-3, while the remaining three originated from the normoxic group (21% O2 CASMCs), denoted as 21% O2 CASMCs-1, 21% O2 CASMCs-2 and 21% O2 CASMCs-3. These libraries underwent high-throughput sequencing analysis, yielding a total of 35.92 g of clean data. Following quality control and data screening procedures, 79,468,573 reads of high quality were obtained. The clean data derived from the six libraries were further processed. High-quality clean reads were aligned to the ribosomal RNA of the target species and subsequently removed, resulting in 77,704,626 valid reads. The average Q30 score was 92.8%, indicating high sequencing accuracy, and the average GC content was 50.013%, consistent with the expected genome distribution. Subsequently, after the quality control of the raw data were compared with the yak reference genome (Bos mutus) (Ensembl_release100, https://ftp.ensembl.org/pub/release-100/gtf/bos_grunniens/), achieving an average alignment rate of 90.584%. This robust alignment rate emphasizes the reliability of the comparison results, affirming their suitability for subsequent data analysis.
Differential expression analysis of DElncRNAs, DEmRNAs and DEmiRNAs
A total of 285 DElncRNAs were identified from the six libraries, of which 212 were up-regulated and 73 were down-regulated (Fig. 4A). These DElncRNAs are visually represented in a heatmap based on their gene expression patterns (Fig. 4B). Additionally, 835 DEmRNAs were identified, of which 608 were up-regulated and 227 were down-regulated (Fig. 4C), and their expression patterns are depicted in a heatmap (Fig. 4D). Furthermore, 126 DEmiRNAs were identified, of which 61 were up-regulated and 65 were down-regulated (Fig. 4E), and their gene expression heatmap is presented (Fig. 4F).
GO enrichment analysis
To further elucidate the functions of DEmRNAs, DElncRNAs and DEmiRNAs in the hypoxic adaptation of yak heart vessels, we conducted GO enrichment analysis of the differential genes using Goatools software. This analysis includes three main aspects: cellular component, molecular function and biological process. As shown in Fig. 5A-D, the top 20 GO terms of DEGs were significantly enriched. Notably, cellular component enrichment primarily involved extracellular matrix components and cellular parts. Molecular function enrichment centered around molecular function regulator, transcription factor activity, protein binding and catalytic activity. Biological process enrichment mainly included immune system processes, metabolic processes and cell development processes.
Enrichment analysis of KEGG signaling pathways
Utilizing the KEGG database, we conducted enrichment analysis to classify the DEmRNAs, DElncRNAs and DEmiRNAs according to the pathways in which they participate. As shown in Fig. 6A-D, the results revealed that the DEGs identified in both the normoxic and hypoxic groups were predominantly enriched in various signaling pathways. These pathways include the TGF-β signaling pathway, MAPK signaling pathway, cAMP signaling pathway, mTOR signaling pathway and PI3K-Akt signaling pathway.
Regulatory ceRNA network of CASMCs in normoxic and hypoxic groups
To gain deeper insight into the regulatory effects of lncRNAs and miRNAs on CASMCs of yak heart under hypoxic and normoxic conditions, we utilized Cytoscape to construct the mRNA-miRNA-lncRNA co-expression network. In this network, mRNAs are represented by green circles, miRNAs by red circles, and lncRNAs by yellow circles. During the analysis, we identified a total of 2660 relationship pairs that met specific criteria (negative correlation ≤ – 0.5, positive correlation ≥ 0.7, and p < 0.05), resulting in numerous relationship pairs that couldn’t be visualized in the network diagram (Fig. 7A). To illustrate, we selected four mRNAs with significant differences and their associated lncRNAs and miRNAs relationship pairs to construct the relevant network graph. For instance, DDIT4 (DNA Damage Inducible Transcrip 4), also known as REDD1, exhibited regulation of DDIT4 expression by 7 miRNAs and 148 lncRNAs (Fig. 7B). Notably, miR-545-x and the lncRNA MSTRG.10328.6, MSTRG.11081.1, MSTRG.1148.1 and MSTRG.11900.1 were involved in this regulatory network. shows that DDIT4 expression is regulated by 31 miRNAs and 66 lncRNAs. miR-545-x and MSTRG.10328.6|MSTRG.11081.1|MSTRG.1148.1|MSTRG.11900.1 displayed the most significant differences. EGLN3, a member of the hypoxia-inducible factor family encoding EGL-9, was found to be regulated by 3 miRNAs and 108 lncRNAs (Fig. 7C). Among them, miR-106-z and the lncRNA MSTRG.10002.1|MSTRG.1148.1|MSTRG.1159.1|MSTRG.11900.1 exhibited the most significant differences. Adenylate activated protein kinase (AMPK), a crucial sensor of intracellular energy regulation, also known as PRKAA2, is regulated by 1 miRNA and 9 lncRNAs (Fig. 7D). Notably, miR-9985-z and the lncRNA MSTRG.10191.1|MSTRG.12914.3|MSTRG.6008.2 were prominent in this regulatory network. TP53INP2, a nuclear localization protein, regulated by 5 miRNAs and 33 lncRNAs (Fig. 7E). Among them, miR-485-x and the lncRNA MSTRG.11450.2|MSTRG.16226.1|MSTRG.16226.2|MSTRG.2032.1 displayed the most significant differences.
The accuracy of the sequencing results was verified by qRT-PCR
To validate the accuracy of sequencing results for DElncRNAs, DEmiRNAs and DEmRNAs, we randomly selected 11 mRNAs, 11 lncRNAs and 8 miRNAs for qRT-PCR analysis. The expression levels of these differential genes were examined in CASMCs of yak hearts under hypoxic and normoxic conditions (Fig. 8A-C). The qRT-PCR results demonstrated that although there were some quantitative differences between the qRT-PCR and RNA-seq analysis platforms, the relative expression levels of DEGs tended to be consistent between the two methods. This consistency demonstrates the reliability of the RNA-seq results, affirming their suitability for bioinformatics analysis.
Discussion
High-altitude adaptation is an extremely complex biological process involving countless molecules and biological processes21. Over time, through natural and artificial selection, yaks have demonstrated remarkable adaptability to harsh and extreme environments, making them an ideal model for studying high-altitude adaptation mechanisms22,23. Previous studies have highlighted the heart as a pivotal organ in yak hypoxia adaptability. In this study, it was found that hypoxia could promote the proliferation of CASMCs in yak heart, suggesting that CASMCs play an important role in the process of hypoxic adaptation in yak heart. To investigate the mechanism, whole transcriptome analysis of yak heart CASMCs in hypoxic and normoxic conditions, yielded a total of 35.92 G of raw data. Additionally, previous studies on the whole transcriptome of yak and bovine heart tissues provided 973,288,574 raw data24,25,26. This highlights the depth and throughput of the sequencing performed on yak heart samples, confirming its validity from a cellular perspective. The average Q20 and Q30 values of filtered bases were 97.33% and 92.8% respectively. Compared with the reference genome, the sequencing coverage was approximately 90.58%, indicating the high accuracy of the high-throughput data obtained in this study.
In this study, we analyzed the expression profiles of DElncRNAs, DEmRNAs and DEmiRNAs to elucidate the genes associated with hypoxic adaptation. Employing criteria of p < 0.5 and fold change ≥ 1.5, we identified a total of 285 DElncRNAs, 835 DEmRNAs and 126 DEmiRNAs in CASMCs under hypoxic and normoxic groups. Notably, this exceeds the number reported in previous studies on miRNA expression profiling in yak and bovine heart tissue20. Subsequently, GO functional enrichment analysis revealed that the differential DEGs between the two groups of cells were primarily enriched in extracellular matrix components, molecular function regulators, immune system processes, metabolic processes, and cell development processes. Previous studies have indicated that numerous genes in high-altitude Tibetan pigs are associated with DNA repair, blood circulation and immunity compared to low-altitude pigs27. Moreover, previous research in our laboratory highlighted the enrichment of immune responses in yak lung tissue, suggesting that the adaptation mechanism of yaks to hypoxic environment without inducing hypoxic damage to lung tissue. Similarly, in this study, we observed that the immune system process in cardiac tissue played an pivotal role in the hypoxic adaptation process of CASMCs. These findings highlight the complex molecular mechanisms underlying hypoxic adaptation in yak heart vessels.
The regulation of signal pathway is crucial in the adaptation of yaks to hypoxia. Through KEGG enrichment analysis, we found that the differential DEGs were primarily involved in in signaling pathways such as HIF-1α, PI3K-Akt, mTOR, MAPK, ECM receptor interaction. Previous studies have revealed that the target genes of yak heart miRNA are predominantly associated with hypoxia-related functions, including HIF-1α, PI3K-Akt and P53 signaling pathways28, which were predominantly associated with previous research, providing initial evidence for the hypoxic adaptation mechanism of CASMCs. Transcriptome analysis of Tibetan chickens and kamara chickens (normoxic control) identified 354 mRNAs, 389 DElncRNAs and 73 DEmiRNAs. It was observed that important miRNAs, along with their target lncRNAs and DEmRNAs, were predominantly involved in angiogenesis and energy metabolism, which is consistent with our findings29. Previous research has underscored the critical role of high energy metabolism levels in yaks during plateau hypoxic adaptation30,31. Additionally, the PI3K-Akt and MAPK signaling pathways are vital for cell proliferation signal transmission, with their interaction being linked to plateau adaptation32,33. Wen used high-throughput sequencing to study the adaptive changes of blood physiology and biochemistry, heart, liver, lung, brain and quadriceps muscle of Tibetan sheep, and found that a large number of hypoxia-related functional genes and pathways could be enriched in the blood pressure indicators and myocardial tissue, resulting in significant changes to adapt to hypoxic changes34. The DEGs identified in this study are not only implicated in the regulation of hypoxia-related pathways, but also may participate in other biological processes. These genes exhibit functions closely associated with cardiovascular development and are involved in pathways releted to heart development and function. Hence, our study posits that these transcriptome factors may exert their effects by modulating the transcriptional expression of target genes during hypoxic adaptation. This regulatory mechanism ensures the normal development of the heart and blood vessels, as well as the proper blood pumping function of plateau yaks, which is conducive to the adaptation to hypoxic environments. By elucidating these regulatory mechanisms, we gain deeper insights into the molecular underpinnings of hypoxic adaptation in yaks, shedding light on potential targets for therapeutic interventions to enhance hypoxia tolerance in various organisms.
In our study, to delve deeper into miRNA regulation, we conducted association analysis of DEmRNAs, DEmiRNAs and DElncRNAs, constructing a comprehensive co-expression network graph of mRNA-miRNA-lncRNA interactions. This network comprised 2660 pairs of relationships, offering valuable insights into the role of yak heart vessels in hypoxic adaptation. Furthermore, combined with the results of this study and the transcriptome studies related to high-altitude hypoxic adaptation such as yak, Tibetan pig, Tibetan chicken and human, we selected DEGs related to vascular system development, hypoxic adaptation and autophagy for qRT-PCR verification, constructing the corresponding co-expression network graph. For instance, the mRNA levels of EGLN3, DDIT4 and BNIP3 were up-regulated, while the mRNA levels of CXCL8, PRKCG, PTGS2, NTF3, AVPR1A, MYLK and TP53INP2were down-regulated. Additionally, certain lncRNAs (MSTRG 16371.3, MSTRG 15951.2, MSTRG 1819.2, MSTRG 2486.1, MSTRG 11450.2, MSTRG 12914.3 and MSTRG 2198.2 ) were down-regulated, while others (MSTRG 7936.12, MSTRG 2010.1, MSTRG 5886.3 and MSTRG 14909.2 ) were up-regulated. Moreover, miR-545-x, miR-19-x, miR-9985-z, miR-106-z and miR-5652-x were down-regulated. miR-709-z, miR-6518-z and miR-1577-z were up-regulated. EGLN3, a member of the hypoxia-inducible factor family encoding EGL-9, lays a crucial role in regulating HIF-1α level under hypoxic conditions, acilitating red blood cell formation and enhancing oxygen transport and utilization by cells35. Similarly, DDIT4, also known as REDD1, was identified as a target gene of HIF-1α in 2002 and was induced to express under hypoxia36. DDIT4 is triggered and expressed under various stress conditions, and its inhibition of the mTOR complex (mTORC1) regulates numerous cellular processes, including protein synthesis, growth, aging, regeneration and autophagy. Studies have revealed that lncRNA C2dat2 promotes autophagy and apoptosis through the miR-30d-5p/DDIT4/mTOR axis in cerebral is chemia/reperfusion, which improves the potential therapeutic target for ischemic stroke treatment37. Indeed, chemokines (CXCL), as a superfamily of chemotactic cytokines, exert pivotal roles in regulating cell migration under inflammatory and physiological conditions38. They facilitate leukocyte mobilization and modulate the immune responses differentiation of recruited cells39, angiogenesis40, organogenesis and germ cell migration41, hypoxia response42and normal development and growth43. Meanwhile, TP53INP2, a nuclear localization protein, shares functional similarities with TP53INP1, which acts as a coactivator of p53 and other transcription factors in the nucleus to regulate the expression of apoptosis and cell cycle-related genes44,45. Notably, study has demonstrated that miR-362-3p exerts a protective effect on cardiomyocyte apoptosis induced by hypoxia/reoxygenation through the inhibition of TP53INP246. In our study, the differentially expressed genes were primarily implicated in signaling pathways such as HIF-1α, PI3K-Akt and autophagy, and fulfilled functions including hypoxic adaptation, angiogenesis and cell proliferation. Moreover, several relationship pairs related to cardiovascular development were enriched, suggesting their significance in the hypoxic adaptation process. Furthermore, we constructed the ceRNA network of related DEGs, providing foundational insights into the specific mechanisms underlying the differential gene functions in hypoxic adaptation.
This study reports for the first time the expression differences of mRNAs, lncRNAs and miRNAs, along with the construction of a differential regulatory network of ncRNAs between the hypoxic and normoxic groups of CASMCs. We identified DDIT4, PRKCG and EGLN3 mas potential key factors for the hypoxic adaptation of the heart and blood vessels, while TP53INP2 emerged as an important regulator of cell autophagy, modulated by ncRNAs. In summary, our findings offer a foundational research direction for exploring the pivotal molecules of ncRNAs influencing the hypoxic adaptation function of yak heart and blood vessels. This study contributes to advancing our understanding of the molecular mechanisms underlying hypoxic adaptation in yaks, paving the way for further investigations aimed at uncovering therapeutic targets for enhancing hypoxia tolerance in various organisms.
Materials and methods
Sample collection
The heart samples of yaks without clinical pathological manifestations were collected from slaughter house (Altitude > 4000 m). Samples were placed for 4 h in sterile saline containing 100 IU/mL penicillin and 100 µg/mL streptomycin and transported back to the laboratory for tissue processing to isolate and culture yak heart CASMCs. All experimental procedures were carried out in accordance with the Code of Animal Ethics of the People’s Republic of China. The study was also approved by the Animal Ethics Committee of Gansu Agricultural University.
The isolation and culture of the yak heart coronary smooth muscle cells
CASMCs were cultured as described previously47,48. Briefly, the adventitia of blood vessels was removed, the isolated coronary arteries were opened, and endothelial cells were gently scraped off with a scalpel blade. The remaining tissues were cut into pieces of 1 mm3 size. The cells were incubated with 1 mg/mL collagenase type I and II in dulbecco’s modified eagles medium (DMEM/F12, 12400024, Gibco, New York, U.S.). The cells were then collected through centrifugation at 1000 rpm for 5 min and resuspended with DMEM/F12 medium supplemented with penicillin (50 IU/mL), streptomycin (50 µg/mL), and fetal bovine serum (20% v/v, A5669701, Gibco). The cell suspension was filtered sequentially with 100, 200, and 400 μm cell strainers. Then, these cells were maintained at 37℃ in a 5% CO2 incubator. When primary cells grew to 90% confluency, cells were digested with 0.25% trypsin containing 0.01% EDTA and then neutralized with DMEM/F12 containing 20% FBS. The cell passage ratio was 1:2 or 1:3, and only 3–5 passages were used in subsequent experiments.
Cell treatment
The experiment was divided into control group and treatment group. In the control group, CASMCs were placed in a standard (37℃, 21% O2, 5% CO2) water jacket cell incubator. To simulate moderate hypoxic conditions, CASMCs were exposed to a three-gas incubator (Thermo Forma 3111, MA, U.S.) (37℃, 5% O2, 5% CO2) that maintained subambient O2 levels by regulated N2 infusion. In all studies, cells were maintained at hypoxic levels throughout the experimental period without any brief reoxygenation periods that could affect the proliferation of CASMCs.
Immunofluorescence analysis
Cells in third passage were cultured to logarithmic growth phase, their density were adjusted to 1 × 104 mL−1, and added to the sterile cover glass in the 6-well plate. After the cells were covered to about 70% of the sterile cover glass, and fixed with 4% PFA for 1 h, incubated for 20 min with 0.5% Triton X-100 for permeabilization, then blocked with 5% bovine serum albumin for 1 h. Then, primary anti-α-SMA (1:100, AF0048, Affinity, Jiangsu, China), anti-Calponin-1 (1:100, bs-0095R, Bioss, Beijing, China), and anti-CD31 (1:200, ab119339, Abcam, Cambridge, UK) antibodies were added and incubated overnight at 4℃. PBS was used as the negative control and anti-rabbit IgG Alexa Fluor 488(1:250, S0018, Affinity) was added for 1 h. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). Digital images were acquired under a fluorescence inverted microscope (Olympus-DP71, Tokyo, Japan).
Cell counting Kit-8 (CCK-8)
In order to study the hypoxia adaptability of yak heart CASMCs, CCK-8 was used to detect the effect of hypoxia on their proliferation activity. To be specific, the cells of 3–4 passages in logarithmic growth phase were added to 96-well plates at a cell density of 3 × 104 mL−1. The plates were placed into the incubator for preculturing for 24 h. The cells were treated with 5% O2 and 21% O2 for 48 h, and then 10 µL of CCK-8 solution and 90 µL of DMEM/F12 medium were added to the well plate and incubated for 2 h. The absorbance was determined at 450 nm using a microplate reader.
Western-blot
In order to study the hypoxia adaptability of yak heart CASMCs, PCNA protein by western blot was detected in yak heart CASMCs treated with normoxic and hypoxic. To be specific, the harvested cells were lysed on ice using RIPA lysis buffer (Solarbio, Beijing, China) for 30 min and protein concentrations were estimated by a BCA Protein Assay Kit (Thermo Fisher Scientific, MA, U.S.). Then, sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed to separate the proteins. The protein was transferred to polyvinylidene fluoride membrane and blocking with 5% skim milk for 2 h. Then, anti-PCNA (1:1000, NBP1-32075, Novus Biologicals, Colorado, U.S.), and β-actin (1:8000, AF7018, Affinity) were added and incubated overnight at 4℃. Next, secondary antibodies goat anti-rabbit (1:5000, HS101, TransGen, Beijing, China) were added and incubated for 1 h. ECL Plus (Vazyme, Nanjing, China) was used to visualize the bands, and the experiment was repeated thrice. Image J software was used to scan the western-blot bands for image analysis, and the relative expression was calculated.
Detection of total RNA quality
Total RNA was isolated from CASMCs treated with samples from yak hearts subjected to normoxic and hypoxic conditions for 48 h using the TRIzol kit following the manufacturer’s instructionsl. RNA quality was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, U.S.) and confirmed via agarose gel electrophoresis.
Construction of lncRNAs and mRNAs library
The remaining RNA was fragmented randomly for cDNA synthesis. A strand-specific method was employed for library construction, involving the use of rRNA-depleted RNA as a template for the first strand cDNA synthesis, followed by the synthesis of the second strand of cDNA using dNTPs, RNase H and DNA polymerase I. The resulting cDNA was purified using AMPure XP Beads, and subsequent steps included cDNA fragment purification, end repair, poly(A) addition, and Illumina HiSeqTM4000 sequencing adapter linkage using the QIAquickPCR extraction kit. The second-strand cDNA was digested with uracil N-glycosylase. Finally, the size selection of cDNA fragments was conducted via agarose gel electrophoresis, followed by PCR, amplification and sequencing by Gene Denovo on the Illumina HiSeqTM4000 platform (Guangzhou, China). Firstly, Fastp (V0.18.0)49was utilized for quality control of the original data, low-quality regions with an unknown nucleotide (N) proportion of less than 10% (> 50% bases, q-value ≤ 20) were removed to yield high-quality data (clean data) for subsequent analysis. The clean reads were then aligned to the ribosome database of the respective species using Bowtie2 (V2.2.8)50. The clean reads were aligned to the reference genome utilizing HISAT2 (V2.1.0)51. Subsequently, the sequences aligned to the reference genome were employed for mRNA assembly using StringTie (V1.3.4) software. The Gene Denovo platform was then utilized for clustering of the differential expression patterns. Expression levels of both lncRNA and mRNA were normalized using FPKM (fragments per kilobase per million reads) to transcript segments. Differential gene coding potential analysis was conducted using CNCI52, CPC253, PFAM54, and CPAT55. In order to more comprehensively excavate the target genes related to the hypoxic adaptive function of yak heart CASMCs, the R package DESeq256 was employed to ascertain the significance of differential lncRNA and mRNA, with a criterion of p < 0.5 and fold change ≥ 1.5 utilized for screening differential genes to identify significant DEGs. It meets the threshold requirements of biological analysis.
Construction of miRNAs library
The small RNA was isolated via polyacrylamide gel electrophoresis (PAGE), selecting bands within the range of 18–30 nucleotides. Subsequently, 3’ and 5’ connectors were individually attached to the RNA. PCR products ranging from 140 to 160 base pairs were enriched to generate cDNA libraries through PCR amplification of reverse transcription products. To ensure data quality, the original data underwent filtering before information analysis. Firstly, raw reads were subjected to quality control using fastp (http://github.com/OpenGene/fastp) to remove low-quality data and obtain clean bases. Reads containing adapter were discarded, and those containing more than 10% of N bases were eliminated. Low-quality reads, defined as those where the number of bases with a quality value Q ≤ 20 accounted for over 50% of the entire read, were also removed. All the reads were then compared with the yak miRNA precursor/mature miRNA sequences in miRBase 21.057 (http://www.mirbase.org/fp.shtml) to determine the observed miRNA sequence and their abundances in the samples. Additionally, the miRNA precursor hairpin structure was utilized to predict new miRNAs. The Milarepa v0.2 (http://sourceforge.net/projects/mireap/) tool was empolyed to analyze sequences without any comments. The yak reference genome and genetic model (Ensembl_release100, https://ftp.ensembl.org/pub/release-100/gtf/bos_grunniens/) were directly download from the the genome website.
Differential expression analysis of DElncRNAs, DEmRNAs and DEmiRNAs
To identify differentially expressed transcripts between various samples or groups, we applied a threshold of fold change ≥ 1.5 and p< 0.05. We utilized DESeq256 and edgeR packages58 for the identification of differentially expressed lncRNAs, miRNAs and mRNAs.
Construction of ceRNA regulatory network
Competitive endogenous RNAs (ceRNAs), as transcriptional regulators, influenced the expression of transcript products through competitive binding. We conducted pathway network analysis to elucidate the function network of differentially expressed mRNAs enriched in KEGG pathway (www.kegg.jp/kegg/kegg1.html). TargetScan and miRanda were employed to predict interactions among differentially expressed mRNA-miRNA-lncRNA trios. The complementary pairs between miRNA and mRNA, miRNA and lncRNA were identified using the hypergeometric test and pearson correlation coefficient, and the ceRNA network were constructed. The standard was set as p < 0.05 and fold change ≥ 1.559,60,61. Visualization of the miRNA-lncRNA-mRNA networks was achieved using Cytoscape (http://cytoscape.org/).
The expression of DElncRNAs, DEmiRNAs and DEmRNAs by qRT-PCR
Primers were designed using Primer Premier 6.0 based on the whole transcriptome sequencing data of CASMCs in both hypoxic and normoxic conditions. A selection of 11 DEmRNAs, 11 DElncRNAs and 8 DEmiRNAs were made from the RNA-seq data based on their potential functional significance for subsequent qRT-PCR validation. The collected cells were used for total RNA extraction by TRIzol method, with their RNA concentration quantified via spectrophotometer. Reverse transcription into cDNA was conducted in a PCR instrument following the steps in the Go Script™ Reverse Transcription System protocol, with resultant cDNA stored at −20℃ for later use. Reverse transcription was performed using Evo M-MLV RT Mix Kit with gDNA Clean for qPCR Kit (Accurate Biotechnology, Hunan, China). Subsequently, SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Hunan, China) was employed in a LightCycler®480 instrument II (Roche, Switzerland). The total reaction volume was 20 µL. The PCR reaction conditions comprised 40 cycles: initial predenaturation at 95℃ for 30 s, denaturation at 95℃ for 5 s, and annealing at 60℃ for 30 s. Melting curve analysis assessed amplification specificity. Comparative Ct method (2−△△Ct) was utilized to calculate the relative expression of transcripts. β-actin served as the reference gene for mRNA and lncRNA, while U6 served as the reference gene for miRNA. Primer specificity was confirmed via BLAST analysis, with single peaks on the melting curve indicating primer specificity. Primer details are provided in Tables 1, 2 and 3.
Statistical analysis
The data are expressed as mean ± standard deviation of three independent experiments. Statistical analyses were conducted using GraphPad Prism 6.0 software (GraphPad software, San Diego, U.S.). One-way analysis of variance was performed using SPSS 21.0 software. nsp ≥ 0.05 indicated nonsignificance, 0.01 ≤ *p < 0.05 denoted significance, and **p < 0.01 signified extreme significance.
Culture characteristics of yak heart coronary smooth muscle cells (CASMCs). (A) Morphological observation of primary CASMCs for 5 and 7 days of in vitro culture. The cultured cells were spindle-like and exhibited a typical “hills and valley” appearance on day 7. Magnification: × 10, Bar = 200 μm. (B) Positive staining for α-SMA and calponin-1, CD-31 exhibited no expression, and the cells were identified as CASMCs. Magnification: × 10, Bar = 200 μm.
Histogram and heat map of DEGs in CASMCs of normoxic and hypoxic groups. (A) Histogram of DElncRNAs in CASMCs of normoxic and hypoxic groups. (B) Heat map of DElncRNAs in CASMCs of normoxic and hypoxic groups. (C) Histogram of DEmRNAs in CASMCs of normoxic and hypoxic groups. (D) Heat map of DEmRNAs in CASMCs of normoxic and hypoxic groups.(E) Histogram of DEmiRNAs in CASMCs of normoxic and hypoxic groups. (F) Heat map of DEmiRNAs in CASMCs of normoxic and hypoxic groups. Red Histogram: Up-regulated genes. Blue Histogram: Down-regulated genes. MNXG 21% O2 : normoxic group. MNXG 5% O2 : hypoxic group.
Histogram and radar chart of GO functional annotation associated with DElncRNAs, DEmRNAs and DEmiRNAs in CASMCs of normoxic and hypoxic groups. (A) DElncRNAs and DEmRNAs related GO functionally annotated antisense histogram in CASMCs of normoxic and hypoxic groups. (B) DElncRNAs and DEmRNAs related GO functionally annotated cis histogram in CASMCs of normoxic and hypoxic groups. (C) DElncRNAs and DEmRNAs related GO functionally annotated trans histogram in CASMCs of normoxic and hypoxic groups. (D) DEmiRNAs related GO functional annotation in CASMCs of normoxic and hypoxic groups. Red bar chart: Biological Process. Green bar chart: Cellular Component. Blue bar chart: Molecular Function.
Bubble plots and radar chart of KEGG pathways associated with DElncRNAs, DEmRNAs and miRNAs in CASMCs of normoxic and hypoxic groups. (A) DElncRNAs and DEmRNAs related KEGG pathways antisense bubble plotsm in CASMCs of normoxic and hypoxic groups. (B) DElncRNAs and DEmRNAs related KEGG pathways cis bubble plotsm in CASMCs of normoxic and hypoxic groups. (C) DElncRNAs and DEmRNAs related KEGG pathways trans bubble plotsm in CASMCs of normoxic and hypoxic groups. (D) DEmiRNAs related KEGG pathway in CASMCs of normoxic and hypoxic groups.
ceRNA network diagram in CASMCs of normoxic and hypoxic groups. (A) ceRNA regulatory network of significantly different genes in CASMCs of normoxic and hypoxic groups. (B) Network diagram of DDIT4 gene in CASMCs of normoxic and hypoxic groups. (C) Network diagram of EGLN3 gene in CASMCs of normoxic and hypoxic groups. (D) Network diagram of PRKAA2 gene in CASMCs of normoxic and hypoxic groups. (E) Network diagram of TP53INP2 gene in CASMCs of normoxic and hypoxic groups.
Comparison of differential gene expression levels of mRNAs, miRNAs and lncRNAs in CASMCs of normoxic and hypoxic groups detected by qRT-PCR. (A) The expression levels of DEmRNAs in CASMCs of normoxic and hypoxic groups. (B) The expression levels of DElncRNAs in CASMCs of normoxic and hypoxic groups. (C) The expression levels of DEmiRNAs in CASMCs of normoxic and hypoxic groups. Green Histogram: hypoxic group. Red Histogram: normoxic group.
Data availability
Raw sequencing reads of lncRNA-seq and miRNA-seq in this paper have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1128418.
References
Zhao, P. F. et al. Different analysis of leptin and its receptor in skin of different parts in young yak (Bos grunniens). J. Agricultural Biotechnol. 31, 530–538 (2023).
Zhang, H. Z. et al. Regulatory effects of HIF-1α and HO-1 in hypoxia-induced proliferation of pulmonary arterial smooth muscle cells in yak. Cell. Signal. 87, 110140 (2021).
Witt, K. E. & Huerta-Sánchez, E. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R Soc. Lond. B Biol. Sci. 374, 20180235 (2019).
Zhou, Y. Y. et al. Effects of intermittent hypoxia on cardiovascular system and its application in the treatment of cardiovascular diseases. Shandong Med. 58, 92–94 (2018).
Wang, Z., Wu, Y. & Wang, Y. Proliferation factors and mechanism of vascular smooth muscle cells. J. Jilin Univ. Med. Ed. 37, 561–566 (2011).
Ma, X. P. Study on anatomical structure of artery and vein in yak heart. Gansu Agricultural Univ. (2008).
He, Y. Y. Changes of the histological structure with age growth and the expression of HIF-1α and VEGF in yak heart. Gansu Agricultural Univ. (2016).
Lee, J. W. et al. Hypoxia signaling in human diseases and therapeutic targets. Exp. Mol. Med. 51, 1–13 (2019).
Gonzalez, F. J., Xie, C. & Jiang, C. The role of hypoxia-inducible factors in metabolic diseasesly. Nat. Rev. Endocrinol. 15, 21–32 (2018).
Weir, E. K. et al. The genetic factor influencing pulmonary hypertension in cattle at high altitude. Cardiovasc. Res. 8, 745–749 (1974).
Hecht, H. H. et al. Brisket disease. III. Clinical features and hemodynamic observations in altitude-dependent right heart failure of cattle. Am. J. Med. 32, 171–183 (1962).
Shao, B. et al. Morphological adaptations of yak (Bos grunniens) tongue to the foraging environment of the Qinghai-Tibetan Plateau. J. Anim. Sci. 88, 2594–2603 (2010).
Wu, T. & Kayser, B. High altitude adaptation in tibetans. High. Alt Med. Biol. 7, 193–208 (2006).
Zhang, X. et al. Report on the physiological and biochemical index for Chinese yak. China Yak. 3, 12–12 (1994).
Xiaoli, K. et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell. Physiol. 234, 9105–9117 (2019).
Paraskevopoulou, M. & Hatzigeorgiou, A. G. Analyzing miRNA–lncRNA interactions. Methods Mol. Biol. (Clifton N J). 1402, 271–286 (2016).
Yao, X. et al. LncRNA ENST00113 promotes proliferation, survival, and migration by activating PI3K/Akt/mTOR signaling pathway in atherosclerosis. Medicine 97, e0473 (2018).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
Wang, K. et al. Different gene expressions between cattle and yak provide insights into high-altitude adaptation. Anim. Genet. 47, 28–35 (2016).
Qi, X. et al. The transcriptomic landscape of yaks reveals molecular pathways for high altitude adaptation. Genome Biol. Evol. 11, 72–85 (2019).
Smith, K. L. M. et al. Chronic developmental hypoxia alters mitochondrial oxidative capacity and reactive oxygen species production in the fetal rat heart in a sex-dependent manner. J. Pineal Res. 73, e12821 (2022).
Menendez-Montes, I. et al. Activation of amino acid metabolic program in cardiac HIF1-alpha-deficient mice. iScience 24, 102124 (2021).
Ayalew, W. et al. Adaptation mechanisms of yak (Bos grunniens) to high-altitude environmental stress. Anim. (Basel). 11, 2344 (2021).
Qiu, Q. G., Zhang, T. & Ma, W. The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949 (2012).
Hu, Q. et al. The yak genome database: an integrative database for studying yak biology and high-altitude adaptation. BMC Genom. 13, 600 (2012).
Wang, H. et al. Whole-transcriptome analysis of yak and cattle heart tissues reveals regulatory pathways associated with high-altitude adaptation. Front. Genet. 12, 579800 (2021).
Li, M. et al. Gcnomic analyses identify distinct patterns of selection in domesticated pigsand tibetan wild boars. Nat. Genet. 45, 1431–1438 (2013).
Ma, J. et al. Comparative transcriptome analysis of Gayal (Bos frontalis), yak (Bos grunniens), and cattle (Bos taurus) reveal the high-altitude adaptation. Front. Genet. 12, 778788 (2022).
Zhang, Y. et al. Comprehensive analysis of coding and non-coding RNA transcriptomes related to hypoxic adaptation in tibetan chickens. J. Anim. Sci. Biotechnol. 12, 60 (2021).
Huang, M. et al. Metabolomics reveals potential plateau adaptability by regulating inflammatory response and oxidative stress-related metabolism and energy metabolism pathways in yak. J. Anim. Sci. Technol. 64, 97–109 (2022).
Wang, H. et al. Comparison of nitrogen metabolism in Yak (Bos grunniens) and indigenous cattle (Bos taurus). J. Anim. Sci. 24, 766–773 (2011).
Ge, Q. Y. et al. Molecular mechanisms detected in yak lung tissue via transcriptome-wide analysis provide insights into adaptation to high altitudes. Sci. Rep. 11, 7786 (2021).
Sang, N. et al. MAPK signaling up-regulates the activity of hypoxia-inducible its effects on p300. J. Biol. Chem. 278, 14013–14019 (2003).
Wen, Y. et al. Changes in the Mitochondrial Dynamics and functions together with the mRNA/miRNA network in the Heart Tissue Contribute to Hypoxic Adaptation in Tibetan Sheep. Anim. (Basel). 12 (5), 583 (2022).
Jaakkola, P. M. & Rantanen, K. The regulation, localization, and functions of oxygen-sensing prolylhydroxylase PHD3. Biol. Chem. 394, 449–457 (2013).
Lv, X. Q. DDIT4 mediates the promoting effect of IL-34 on the proliferation in monocytic leukemia cells. Peking Union Med. Coll. (2022).
Xu, Q. et al. lncRNA C2dat2 facilitates autophagy and apoptosis via the miR-30d-5p/DDIT4/mTOR axis in cerebral ischemia-reperfusion injury. Aging (Albany NY). 13, 11315–11335 (2021).
Zlotnik, A. & Yoshie, O. Chemokines: a new classification system and their role in immunity. Immunity 12, 121–127 (2000).
Alejo, A. & Tafalla, C. Chemokines in teleost fish species. Dev. Comp. Immunol. 35, 1215–1222 (2011).
Keane, M. et al. CXC chemokines and angiogenesis/angiostasis. Proc. Assoc. Am. Physicians. 110, 288–296 (1998).
Doitsidou, M. et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111, 647–659 (2002).
Schioppa, T. et al. Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 198, 1391–1402 (2003).
Zeng, Q. et al. Transcriptome display during testicular differentiation of channel catfish (ictalurus punctatus) as revealed by RNA-Seq analysis. Biol. Reprod. 95, 19 (2016).
Okamura, S. et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell. 8, 85–94 (2001).
Tomasini, R. et al. TP53INP1 is a novel p73target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene 24, 8093–8104 (2005).
Shi, J. et al. MicroRNA-362-3p implicated in cardioprotection against hypoxia/ reoxygenation-induced cardiomyocyte apoptosis by repressing TP53INP2 and regulating SDF-1/CXCR4 pathway. Altern. Ther. Health Med. 29, 254–261 (2023).
Husarek, K. E. et al. Isolation of murine coronary vascular smooth muscle cells. J. Vis. Exp. 111, 53983 (2016).
Montezano, A. C. et al. Touyz RM. Isolation and culture of vascular smooth muscle cells from small and large vessels. Methods Mol. Biol. 1527, 349–354 (2017).
Chen, S. et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 9, 357–359 (2012).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Sun, L. et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41, e166 (2013).
Kong, L. et al. Cpc: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349 (2007).
Finn, R. et al. Pfam: the protein families database. Nucleic Acids res. 42, D222–D230 (2014).
Wang, L. et al. Cpat: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Griffiths-Jones, S. et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 (2006).
Robinson, M. D., Mccarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for diferential expression analysis of digital gene expression data. Biogeosciences 26, 139–140 (2010).
Wang, J. et al. Identifcation of the regulatory role of lncRNA SNHG16 in myasthenia gravis by constructing a competing endogenous RNA network. Mol. Ther. Nucleic Acids. 19, 1123–1233 (2020).
Liang, R. et al. Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 7, 6396 (2017).
Zuo, J. et al. Analysis of the coding and non-coding RNA transcriptomes in response to bell pepper chilling. Int. J. Mol. Sci. 19, 2001 (2018).
Acknowledgements
We thank the local commercial slaughterhouse in Xining City, Qinghai Province for providing the experimental animals.
Funding
This work was supported by the National Natural Science Foundation of China (Grants No. 32372974).
Author information
Authors and Affiliations
Contributions
S.S.Y. and Y.C conceived the study. S.S.Y. and R.M. collected the samples. S.S.Y., X.F.B. and H.Z. carried out the experiment. S.S.Y. and P.F.Z. analyzed the data. S.S.Y. wrote the manuscript. S.S.Y., Y.C., S.J.Y. and J.F.H reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All experimental procedures were handled according to the Animal Ethics Procedures and Guidelines ofthe People’s Republic of China. The study was also approved by the Animal Ethics Committee of GansuAgricultural University (GSAU-Eth-VMC-2023-004).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, S., Cui, Y., Yu, S. et al. Integrated analysis of the expression profiles of the lncRNA-miRNA-mRNA ceRNA network in CASMCs under hypoxia and normoxia conditions in yak heart. Sci Rep 15, 9165 (2025). https://doi.org/10.1038/s41598-025-85483-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-85483-4