Abstract
Deep learning-based medical image segmentation methods are generally divided into convolutional neural networks (CNNs) and Transformer-based models. Traditional CNNs are limited by their receptive field, making it challenging to capture long-range dependencies. While Transformers excel at modeling global information, their high computational complexity restricts their practical application in clinical scenarios. To address these limitations, this study introduces VMAXL-UNet, a novel segmentation network that integrates Structured State Space Models (SSM) and lightweight LSTMs (xLSTM). The network incorporates Visual State Space (VSS) and ViL modules in the encoder to efficiently fuse local boundary details with global semantic context. The VSS module leverages SSM to capture long-range dependencies and extract critical features from distant regions. Meanwhile, the ViL module employs a gating mechanism to enhance the integration of local and global features, thereby improving segmentation accuracy and robustness. Experiments on datasets such as ISIC17, ISIC18, CVC-ClinicDB, and Kvasir demonstrate that VMAXL-UNet significantly outperforms traditional CNNs and Transformer-based models in capturing lesion boundaries and their distant correlations. These results highlight the model’s superior performance and provide a promising approach for efficient segmentation in complex medical imaging scenarios.
Similar content being viewed by others
Introduction
High-resolution medical images are crucial in modern medicine, but their complexity and diversity pose significant challenges to traditional segmentation methods1. For instance, retinal vessel segmentation often suffers from low contrast, high noise, and brightness variations, leading to insufficient segmentation accuracy. Additionally, the vast amount of data further increases the time and cost of manual annotation.
With the rapid development of computer and artificial intelligence technologies, convolutional neural networks (CNNs) have demonstrated powerful modeling capabilities and gained widespread attention. Among them, UNet2, a deep learning architecture designed specifically for biomedical image segmentation, features a symmetric “U”-shaped structure comprising an encoder and a decoder. This design enables UNet to achieve outstanding performance in handling the complex structures in medical images. Following this direction, various UNet variants such as UNet++3, U-Net V24, and UNet 3+5 have been developed and successfully applied to image and volume segmentation across diverse medical imaging modalities. However, due to the inherent limitations of convolution operations, CNN-based networks struggle to effectively capture global context and long-range dependencies, limiting their performance in complex semantic modeling tasks.
To overcome these limitations, researchers have drawn inspiration from the success of Transformers in natural language processing and introduced Transformer-based models into computer vision. Vision Transformer (ViT)6, one of the first models fully based on multi-head self-attention, efficiently captures long-range dependencies and encodes complex shape information, demonstrating remarkable modeling capabilities. Building on this, improved models such as Swin Transformer7, which incorporates local window self-attention to reduce computational complexity, and DeiT8, which optimizes training strategies for smaller datasets, have been proposed. While these models achieve notable performance improvements, pure Transformer architectures remain limited in capturing local details and suffer from high computational complexity, especially when processing high-resolution images, posing challenges for clinical applications. To address these issues, hybrid models combining CNNs and Transformers have been explored to enhance network performance. For example, TransUNet9 integrates Transformers with the UNet architecture, significantly improving medical image segmentation by enhancing global feature extraction. However, these methods have yet to strike an ideal balance between improving performance and reducing computational cost, necessitating further optimization.
Recently, Mamba10 has garnered attention for its powerful sequence modeling capabilities. Particularly in medical image segmentation, integrating Mamba into classic UNet architectures effectively captures long-range dependencies. As a structured state space model (SSM), Mamba offers linear computational complexity and efficient global feature modeling, making it an ideal solution for processing global context information in segmentation models. Similarly, the recently proposed xLSTM11, an extended version of traditional LSTM, enhances long-range dependency modeling through optimized gating mechanisms. Compared to Transformers, xLSTM not only achieves linear computational complexity but also excels in capturing sequence details. Although initially applied to natural language processing and image classification, xLSTM’s potential in medical image segmentation is worth exploring.
The key to achieving accurate medical image segmentation lies in effectively capturing and integrating local features and long-range correlations. While combining CNNs and Transformers offers a promising direction, alternative approaches may also provide effective solutions. Inspired by VMamba12 and xLSTM, this paper proposes VMAXL-UNet, a novel segmentation model designed to overcome the limitations of existing methods and further improve segmentation accuracy.
VMAXL-UNet inherits the classic UNet design elements, such as the encoder-decoder architecture and skip connections, while integrating the strengths of SSM and xLSTM to enhance its ability to model complex dynamic systems and long-range dependencies. To further boost performance, VMAXL-UNet employs a four-layer encoder structure, where the first three layers consist of VSS blocks and BasicConv blocks, and the fourth layer combines VSS blocks with ViL blocks, incorporating patch merging for down-sampling to enhance feature extraction. The decoder mirrors this design with four layers, each composed of two BasicConv blocks, and employs patch expansion for up-sampling to restore the segmentation output’s resolution. Skip connections integrate features via additive operations, further improving segmentation performance.
Extensive experiments were conducted on multiple medical image datasets, including ISIC1713, ISIC1814, Kvasir-SEG15, and ClinicDB16, to validate the proposed model. The results demonstrate that VMAXL-UNet achieves superior segmentation performance across these datasets, showcasing its strong capability and potential in medical image segmentation.The primary contributions of this work are as follows:
-
1.
Introduction of the ViL module for medical image segmentation, enhancing the model’s ability to handle complex lesion morphology and blurred boundaries, thereby improving segmentation accuracy, especially in fine-structure recognition and boundary clarity.
-
2.
Proposal of VMAXL-UNet, an encoder-decoder architecture combining SSM and xLSTM, tailored for medical image segmentation tasks.
-
3.
Comprehensive experiments conducted on four datasets, demonstrating the competitive performance and broad applicability of VMAXL-UNet in medical image segmentation.
Related Work
Applications of CNNs and transformers in medical image segmentation
Over the past few decades, Convolutional Neural Networks (CNNs) have been a core component of artificial neural networks, achieving remarkable success in deep learning and computer vision, and have found widespread application in the medical imaging field. Medical image segmentation, as a crucial task in image processing, witnessed a historic breakthrough with the introduction of U-Net. U-Net effectively performs pixel-level classification through its encoder-decoder symmetric structure, significantly improving segmentation accuracy. Inspired by U-Net, UNet++3 was developed by incorporating dense skip connections to address the semantic gap during feature fusion. Subsequent research introduced techniques such as attention mechanisms17, image pyramids18, and residual networks19, further enhancing the performance of CNN-based segmentation methods. The Transformer architecture, initially proposed by Vaswani et al.20 in 2017 for natural language processing, gained significant attention for its superior ability to handle long-range dependencies and complex contextual relationships. In 2020, Dosovitskiy et al.6 introduced the Vision Transformer, applying the Transformer architecture to image classification tasks in computer vision. Since then, Transformer-based methods for medical image segmentation have emerged, with notable works such as Swin-Unet21, which utilizes the Swin Transformer to enhance feature representation, marking the first Transformer-based U-Net architecture. Although both CNNs and Transformers have demonstrated impressive capabilities in image segmentation tasks, each has its limitations. As a result, many studies have begun to explore ways to combine the strengths of both CNNs and Transformers. For instance, UCTransNet22 replaces the skip connections in U-Net with Transformer-based modules to enhance the fusion of global and local features. However, despite progress in the fusion of local and global features, these methods still face challenges in meeting the high-accuracy segmentation requirements in medical imaging.
Applications of Mamba and LSTM in medical image segmentation
In recent years, State Space Models (SSMs) have been introduced into deep learning for sequence modeling, with their parameters or mappings learned through gradient descent23. SSMs essentially serve as a sequence transformation method that can be effectively integrated into deep neural networks. However, due to the computational and storage demands of state representations, SSMs have not been widely adopted in practical applications. This situation changed with the introduction of the Structured State Space Model (S4), which addresses the computational and storage limitations of traditional SSMs by reparameterizing the state matrix24. Subsequently, Mamba10, one of the most successful variants of SSMs, emerged, significantly enhancing the application of SSMs. Mamba not only retains modeling performance comparable to Transformers but also exhibits linear scalability, enabling it to efficiently handle long sequence data. This makes SSMs a strong competitor to Transformers. The advantages of Mamba have led to its rapid prominence in sequence modeling and demonstrated substantial potential in medical image processing, especially when combined with U-Net, further improving medical image segmentation accuracy. For example, models like Mamba-UNet25 and VM-UNet26 introduced the Visual Mamba module, constructing U-Net-like architectures that significantly enhanced multi-scale feature extraction capabilities, optimizing segmentation performance. The successful application of these models not only highlights Mamba’s potential in medical image segmentation but also provides new insights for the further development of deep learning models in medical image processing.
Long Short-Term Memory (LSTM) networks27, proposed by Hochreiter and Schmidhuber in 1997, were designed to address the vanishing gradient problem encountered by traditional Recurrent Neural Networks (RNNs) when processing long sequence data. LSTMs are capable of effectively processing sequential data while retaining information from earlier steps in the sequence, making them particularly well-suited for tasks involving long-term dependencies28. This ability allows LSTMs to learn and remember long-term dependencies, where information from earlier time steps is crucial for predicting later steps. In the field of medical imaging, LSTMs have gradually gained significant attention and application. For example, Salehin29proposed an LSTM-based method for medical image classification, MedvLSTM, which shows great potential in improving the accuracy and efficiency of medical image classification. Shahzadi et al.30, addressing the limitations of traditional Convolutional Neural Networks (CNNs) in 3D medical image classification, particularly the challenges in optimizing 3D volume classification tasks, proposed a cascaded model combining CNNs and LSTMs, CNN-LSTM, which was applied to classify brain tumor MRI images and effectively distinguish between high-grade and low-grade gliomas. Despite the success of LSTM in medical image processing, its inherent limitations, such as limited storage capacity, inability to correct storage decisions, and lack of parallel processing capability, still constrain its further application. To overcome these issues, xLSTM11 was introduced, addressing the shortcomings of traditional LSTMs in terms of flexibility in information storage and parallel processing of long sequences, breathing new life into LSTMs in modern AI applications. For example, the xLSTM-UNet31 model, by incorporating xLSTM into the U-Net architecture, enhanced the model’s ability to capture sequential data and significantly improved medical image segmentation accuracy. This innovative combination not only optimized LSTM performance but also boosted the accuracy and efficiency of medical image analysis.
Methods
Architecture overview
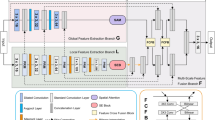
The overall architecture of the model is shown in Fig. 1, which consists of three main components: the encoder, decoder, and skip connections. In the encoder, the input medical image is first divided into non-overlapping patches of size 4\(\times\)4, transforming the input into a sequence of embeddings. The input image has dimensions of H\(\times\)W\(\times\)3, and after passing through the patch embedding layer, the image is mapped to C channels, resulting in an embedded image of size H/4 \(\times\) W/4 \(\times\) C. This embedded image is then passed into the encoder for feature extraction. The encoder consists of four stages, with patch merging operations applied at the end of the first three stages to gradually reduce spatial resolution and increase the number of channels, thereby enhancing the model’s ability to extract image features. Specifically, the first three stages are composed of VSS blocks and BasicConv blocks, which help capture both local features and long-range dependencies in the image. The final stage consists of VSS and ViL blocks, further improving feature representation and strengthening the modeling of long-range dependencies. The number of channels at each stage is [C, 2C, 4C, 8C], enabling the model to effectively extract and fuse features at different levels.
The decoder is also divided into four stages. In the last three stages, patch expanding operations are applied, gradually reducing the number of feature channels and increasing the spatial resolution of the feature maps, thereby restoring image details. The number of channels in the BasicConv blocks used in the decoder stages is [8C, 4C, 2C, C], and the number of BasicConv layers at each stage is [2, 2, 2, 2]. This design helps the decoder progressively restore the image’s spatial resolution, ensuring that the output dimensions match the input image’s size before the final output. After the decoder, the Final Projection layer adjusts the feature dimensions to the appropriate size for the segmentation task, ultimately generating the segmentation result. Skip connections play a critical role throughout the model by directly fusing features between the encoder and decoder through addition, which helps retain more detailed information and, consequently, improves segmentation accuracy.
VSS block
The VSS block is derived from VMamba12, and its module structure is illustrated in Fig. 1. In the VSS block, the input data is first processed by Layer Normalization and then split into two streams. In the first stream, the data passes through a linear layer followed by a nonlinear transformation via an activation function. In the second stream, the data is also processed through a linear layer, followed by a 3\(\times\)3 convolutional layer and an activation function to further extract local features. The output from the second stream is then fed into the 2D Selective Scanning (SS2D) module for deeper feature extraction. The features processed by the SS2D module undergo Layer Normalization to ensure consistency in the feature distribution. The normalized features are then element-wise multiplied with the output of the first stream, merging the information from both paths. Finally, the merged features are passed through a linear layer and combined with the original input data via a residual connection, forming the final output of the VSS block.
The SS2D module consists of three components: the scanning expansion operation, the S6 module, and the scanning merging operation. The scanning expansion operation, as shown in Fig. 2a, unfolds the input image into sequences along four different directions. These sequences are then processed by the S6 module for feature extraction, ensuring that information from all directions is fully scanned, thereby capturing diverse features. Subsequently, the scanning merging operation, as depicted in Fig. 2b, sums and merges the sequences from the four directions, restoring the output image to the same size as the input image. Specifically, given the input feature \(w\), the output feature \(\bar{w}\) of SS2D can be expressed as:
where \(z \in V = \{1, 2, 3, 4\}\) represents the four different scanning directions (as shown in Figure 2), \(\text {Expand}(\cdot )\) and \(\text {Merge}(\cdot )\) correspond to the scanning expansion and scanning merging operations, respectively, and \(S6(\cdot )\) denotes the output after passing through the S6 module. The Selective Scanning State Space Sequence Model (S6) in Equation (2) is the core of the VSS block, responsible for processing the input sequence through a series of linear transformations and discretization processes. For more details on S6, please refer to10.
ViL Block
For the ViL module (as shown in Fig. 1), the input information first undergoes Layer Normalization to standardize the data distribution and stabilize the training process. The input is then split into two streams. In the first stream, the data is passed through a linear layer for linear transformation, followed by a SiLU activation function for nonlinear processing, enhancing the model’s expressive power. In the second stream, the data is also processed through a linear layer, followed by a convolutional layer to capture local features, with an activation function further enriching the feature representations. Subsequently, the output from the second stream is fed into the mLSTM module to capture long-range dependencies and further extract features. After processing by the mLSTM module, the extracted features undergo Layer Normalization again to ensure consistency in feature distribution. The normalized features are then element-wise multiplied with the output from the first stream, effectively integrating the information from both paths. Finally, the merged features are processed through a linear layer and combined with the original input data via a residual connection, resulting in the final output of the ViL module. This design not only captures local features but also effectively models long-range dependencies, thereby enhancing the overall feature representation capability.
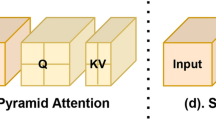
mLSTM (Matrix LSTM), derived from xLSTM11, significantly enhances the model’s memory and parallel processing capabilities by extending the vector operations in traditional LSTM to matrix operations. In mLSTM, each state is no longer represented by a single vector but by a matrix. This design allows it to capture more complex data relationships and patterns within a single time step. Additionally, mLSTM employs the FlashAttention mechanism, which dynamically guides the updating process of cell states and normalized states through the interaction of queries, keys, and values, ultimately generating the final hidden layer output. This design not only improves the model’s ability to model complex data patterns but also significantly boosts computational efficiency, as illustrated in Fig. 3. Specifically, the mLSTM layer first performs linear projections on the query, key, and value vectors:
where \(x_t\) denotes the input vector, \(W_q\), \(W_k\), and \(W_v\) are the corresponding mapping (or weight) matrices, and \(b_q\), \(b_k\), and \(b_v\) are the corresponding bias terms.
mLSTM uses input gates and forget gates to control memory updates, and employs exponential gating and OR gating (OR gating) to facilitate matrix memory computations:
where \(w_i^T\), \(w_f^T\), \(b_i\), and \(b_f\) denote the weight vectors and bias terms corresponding to the input gate and forget gate, respectively. \(\sigma\) represents the activation function, and \(\exp (\cdot )\) denotes the exponential operation.
mLSTM extends the memory cell to a matrix and combines the update mechanism of LSTM with the information retrieval scheme from Transformer, introducing an integrated attention-based cell state and hidden state update mechanism, allowing memory extraction from different time steps:
The cell state is updated using a weighted sum according to the ratio, where the forget gate corresponds to the weighted proportion of memory, and the input gate corresponds to the weighted proportion of key-value pairs, satisfying the covariance-based update rule. mLSTM adopts a normalizer that weights the key vectors. Finally, through normalization and weighted processing controlled by the output gate, the network’s hidden state \(h_t\) is obtained.
For more details on mLSTM, please refer to11.
BasicConv block
Each basic residual block consists of a convolutional layer (Conv), instance normalization (Instance Normalization), and a ReLU activation function, as shown in Fig. 1. This paper uses a \(3 \times 3\) convolution kernel, stride of 1, and padding of 1. Each sub-module adopts residual connections, adding the input directly to the output, thereby forming residual learning. Formally, given an input image \(W_0 = I \in \mathbb {R}^{H_0 \times W_0 \times C_0}\), the final output is:
where \(Y_1\) represents the output of the first submodule, \(Y_2\) is the output feature map of the second submodule, \(\text {Conv}(\cdot )\) denotes the convolution operation output, \(\text {IN}(\cdot )\) indicates the Instance Normalization operation, and \(\text {RELU}(\cdot )\) refers to the Rectified Linear Unit (ReLU) activation function.
In the decoder, the upsampled feature maps can undergo convolution operations, with residual connections preserving the input features. This process further refines the feature information, aids in recovering high-quality details, and helps mitigate the potential vanishing gradient problem during information propagation.
Encoder
Given an input image \(Z_0 \in \mathbb {R}^{H \times W \times 3}\), the encoder progressively compresses the input image information through multi-level feature extraction and downsampling operations. It achieves efficient fusion of global and local information via the VSS and ViL modules. Specifically, the VSS module utilizes the Structured State Space Model (SSM) for global information modeling, and incorporates depthwise separable convolutions (DW-CNN) to further optimize the features. The output of the VSS block is then given by:
where Z represents the input image, \(DWConv(\cdot )\) denotes the output of the depthwise separable convolution (DW-CNN) operation, \(SS2D(\cdot )\) represents the output after passing through the 2D selective scanning (SS2D) module, and \(\text {Linear}(\cdot )\) indicates a linear transformation operation used to map and reorganize the channel dimension.
The ViL module enhances the modeling ability of local details and edge information through mLSTM, making it particularly suitable for capturing lesion boundary information. The input data is first normalized as follows:
The features from the two paths are fused as follows:
Finally, the output of the ViL block is:
where \(Z\) represents the input image, \(\text {LayerNorm}(\cdot )\) denotes the normalization of feature maps, \(\text {SiLU}(\cdot )\) represents the nonlinear activation function, and \(\text {Linear}(\cdot )\) refers to the linear transformation operation applied to map and reorganize the channel dimensions. The symbol \(\odot\) denotes element-wise multiplication, \(\text {Conv}(\cdot )\) represents the output of the convolution operation, and \(\text {mLSTM}(\cdot )\) represents the output after processing through the mLSTM module.
In the final layer of the encoder, a gating mechanism is employed to adaptively fuse the VSS and ViL features, thereby balancing global and local information. The gating mechanism is defined as:
where \(\sigma\) is the Sigmoid function, and \(W_g\) and \(b_g\) are learnable parameters. Thus, the fused feature representation is given by:
where \(\odot\) denotes element-wise multiplication.
Decoder
The decoder progressively restores the multi-level features extracted by the encoder to the original image resolution through upsampling and skip connections, enabling precise segmentation. Each layer of the decoder receives multi-scale features from the encoder through skip connections, enhancing the model’s ability to recover segmentation details. The effective transmission and reconstruction of features are ensured through the BasicConv block:
where \(F_{\text {skip}}\) represents the skip connection feature map from the encoder, \(\text {PatchExpand}(\cdot )\) denotes the upsampling operation applied to the input feature map to increase its spatial resolution, \(F_{\text {up}}\) is the upsampled feature map from the previous decoder layer, and \(\text {BasicConv}(\cdot )\) represents the output after passing through the BasicConv block.
Experiments and results
Datasets
ISIC dataset
In this study, we selected two skin lesion segmentation datasets, namely ISIC1713 and ISIC1814. These datasets contain a large number of high-quality annotated skin lesion images, with 2150 and 2694 labeled images, respectively. We split the data into training and testing sets at a ratio of 7:3. Specifically, for the ISIC17 dataset, 1505 images were used for training, and 645 images were used for testing. For the ISIC18 dataset, 1886 images were allocated to the training set, while 808 images were used for testing.
Polyp segmentation dataset
For the polyp segmentation task, we utilized two common endoscopic image datasets, Kvasir-SEG15 and ClinicDB16. These datasets consist of high-definition endoscopic images primarily obtained from colonoscopy and gastroscopy procedures. The Kvasir-SEG dataset contains 1000 labeled images, while the ClinicDB dataset includes 612 labeled images. Following the experimental setup of PraNet32, we adopted a separate training and testing strategy: 900 images from the Kvasir-SEG dataset and 550 images from the ClinicDB dataset were used for training, with the remaining images allocated to the testing set.
Experimental details
The experiment was carried out on an Ubuntu 22.04 system, utilizing an environment equipped with Python 3.10.4, PyTorch 2.3.0, and CUDA 11.8. All experimental tasks were executed on a single NVIDIA A10 GPU.We resized all images in the datasets to 256\(\times\)256 pixels and employed data augmentation techniques such as random flipping and random rotation to prevent overfitting. Regarding operational parameters, the batch size was set to 32, the optimizer used was AdamW33, with an initial learning rate of 2.3e-4. We also utilized CosineAnnealingLR34 as the learning rate scheduler, with a maximum of 50 epochs, setting the minimum learning rate to 1e-5, and the training cycles were set to 500 iterations. During model training, the weights of the encoder were initialized using VMamba-S12 weights pretrained on ImageNet-1k.
Loss function
Based on the characteristics of binary cross-entropy and the Dice similarity coefficient, and considering that all our dataset masks consist of two classes (target and background), we designed a hybrid loss function with \(\beta _1 = 1\) and \(\beta _2 = 1\). This ensures that the loss function effectively distinguishes between the target and background while maintaining a balanced treatment of both classes. The formula is as follows:
Where N represents the total number of samples, \(y_i\) and \(p_i\) denote the true label and the predicted value for pixel i, respectively. \(L_{\text {Bce}}\) represents the binary cross-entropy loss, which measures the difference between the model’s predictions and the true labels. \(L_{\text {Dice}}\) represents the Dice loss, used to assess the overlap between the model’s predicted segmentation and the true label. \(\beta _1\) and \(\beta _2\) are two weighting factors that control the relative importance of the binary cross-entropy loss (\(L_{\text {Bce}}\)) and the Dice loss (\(L_{\text {Dice}}\)).
Results
We compare the proposed VMAXL-UNet model with several state-of-the-art methods, including CNN-based UNet and EGEUNet models, Transformer-based Swin-Unet, Mamba-based VM-UNet, the recently proposed xLSTM-based xLSTM-UNet, and MLP-based U-KAN. In the experiments, we use Intersection over Union (IoU) and Dice coefficient as evaluation metrics. The evaluation results on the ISIC dataset are shown in Table 1, while the results on the selected Polyp segmentation datasets are presented in Table 2. The results demonstrate that our VMAXL-UNet model outperforms other methods across all four datasets. Notably, on the Kvasir-SEG and ClinicDB datasets, VMAXL-UNet achieves significant advantages. These two datasets contain many targets with blurry boundaries, making it difficult to distinguish them from the background, which highlights VMAXL-UNet’s ability to effectively capture long-range dependencies to enhance segmentation performance. In addition to the accuracy advantage, we further demonstrate the efficiency of our model as a network baseline. As shown in Table 3, we present the computational complexity (FLOPs) and the number of parameters (Params), along with the average segmentation accuracy, across the four datasets. The experimental results indicate that our proposed model not only surpasses most segmentation methods in terms of accuracy but also demonstrates considerable efficiency. Overall, in the trade-off between segmentation accuracy and efficiency, our method exhibits the best performance.
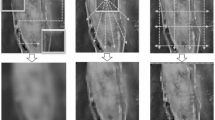
To further validate the segmentation performance of our model, we conducted a visual analysis of the segmentation results on the ISIC17, ISIC18, Kvasir-SEG, and ClinicDB datasets, as shown in Figs. 4 and 5. From the visual results, it is evident that the proposed VMAXL-UNet model significantly outperforms the other comparative models in terms of segmentation quality. Even when processing small objects, VMAXL-UNet not only accurately localizes the target regions but also generates coherent and clear boundaries. These results strongly demonstrate that VMAXL-UNet excels in both local feature extraction and global context modeling. The model’s outstanding performance in fine-grained segmentation tasks further validates its practicality and robustness in complex medical image segmentation scenarios.
Ablation studies
To investigate the impact of various factors on model performance and to validate the effectiveness of the proposed model, a comprehensive ablation study was conducted on the ISIC17 dataset. As shown in Tables 4 and 5, two experiments were designed and implemented.
In the first experiment, we modified the encoder part of the model and constructed three different variant models: Model 1 consists only of BasicConv blocks; Model 2 is composed of BasicConv and VSS blocks; Model 3 represents the original VMAXL-UNet model. The experimental results indicate that, although Model 1, which only uses BasicConv blocks, outperforms the traditional U-Net model, demonstrating the inherent advantage of the BasicConv block, further incorporation of the VSS and ViL modules (i.e., Model 3) significantly enhances overall performance, particularly achieving the best segmentation accuracy. This result fully confirms the substantial contribution of the VSS and ViL modules to model performance.
In the second experiment, we further explored the impact of the number of VSS+ViL module blocks in the encoder on model performance. Three scenarios were tested, with the number of VSS+ViL blocks set to 0, 1, and 2, respectively. When the number of VSS+ViL blocks was 0, the encoder was composed solely of BasicConv and VSS blocks. The experimental results showed that the model performance did not increase monotonically with the number of VSS+ViL blocks. Specifically, when the number of VSS+ViL blocks was 1, the model achieved higher segmentation accuracy, indicating that a moderate number of VSS+ViL blocks can effectively enhance model performance.
The results of these experiments not only validate the importance of the VSS and ViL modules but also provide valuable insights for determining the optimal configuration of module quantities in model design.
Attention visualization for comparative analysis
To further validate the design rationale and practical application potential of VMAXL-UNet, we present the attention heatmaps of VMAXL-UNet and its two variant models (Model 1 and Model 2) in the medical image segmentation task in Fig. 6. These heatmaps visually reflect the model’s attention to key regions in the input images. The input images contain complex intestinal structures and lesion areas, indicating a high level of difficulty in the segmentation task; the ground truth represents manually annotated lesion regions, serving as the standard reference for segmentation results. The high-response regions (indicated in red) of VMAXL-UNet precisely cover the lesion areas, showing a high degree of consistency with the ground truth. This is attributed to the integration of the SSM and xLSTM modules in the encoder: the former enhances the model’s attention to detail regions by efficiently fusing multi-scale features, while the latter improves the model’s ability to capture global context information. In contrast, the high-response regions of the variant Model 1 (which does not integrate the SSM and xLSTM modules in the encoder, instead using BasicConv blocks) exhibit some diffusion, making it difficult to accurately focus on the lesion boundaries, potentially leading to blurry boundaries in the segmentation results. The attention distribution of variant Model 2 (which integrates only the SSM module in the encoder) is more dispersed, with some high-response areas deviating from the lesion regions, indicating insufficient adaptability in complex scenarios. These comparative results demonstrate that VMAXL-UNet outperforms the variant models in capturing lesion details and boundary information, thereby fully validating its design effectiveness and practical potential in medical image segmentation tasks.
Conclusions
This study presents VMAXL-UNet, a novel UNet variant based on the State Space Model (SSM) and an enhanced Long Short-Term Memory network (xLSTM). By introducing the Visual State Space (VSS) module and the ViL module, VMAXL-UNet demonstrates significant performance improvements in medical image segmentation tasks. Specifically, the VSS module leverages visual saliency analysis to enable the model to focus more effectively on critical features, while the ViL module enhances the modeling of sequential dependencies in complex image structures. Additionally, the incorporation of VMamba pre-trained weights accelerates model convergence and improves initial performance.
Experiments on several medical image datasets, such as dermatological and polyp segmentation tasks, demonstrate that VMAXL-UNet outperforms or matches existing state-of-the-art models across multiple evaluation metrics. However, there is still room for optimization in terms of computational efficiency. Future work could explore more lightweight network architectures or improved training strategies to reduce computational costs. Additionally, the design of VMAXL-UNet is highly versatile, and further research will investigate its potential in 3D medical image segmentation and other image processing tasks, such as organ segmentation and tumor detection, to assess its applicability and generalization capabilities.
Data availability
The data used in this study is publicly accessible, with ISIC17 and ISIC18 datasets sourced from the ISIC Challenge. The access link is https://challenge.isic-archive.com/data/. For the Kvasir-SEG and ClinicDB datasets, they can be accessed through their official websites: https://datasets.simula.no/kvasir-seg/ and https://opendatalab.org.cn/OpenDataLab/CVC-ClinicDB, respectively.
References
Muksimova, S., Umirzakova, S., Mardieva, S. & Cho, Y.-I. Enhancing medical image denoising with innovative teacher-student model-based approaches for precision diagnostics. Sensors 23, 9502 (2023).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical image computing and computer-assisted intervention-MICCAI 2015: 18th international conference, Munich, Germany, October 5–9, 2015, proceedings, part III 18 (ed. Ronneberger, O.) 234–241 (Springer, 2015).
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4 (ed. Zhou, Z.) 3–11 (Springer, 2018).
Peng, Y., Sonka, M. & Chen, D.Z. U-net v2: Rethinking the skip connections of u-net for medical image segmentation. Preprint at arXiv:2311.17791 (2023).
Huang, H. et al. Unet 3+: A full-scale connected unet for medical image segmentation. In ICASSP 2020–2020 IEEE international conference on acoustics, speech and signal processing (ICASSP) (ed. Huang, H.) 1055–1059 (IEEE, 2020).
Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. Preprint at arXiv:2010.11929 (2020).
Liu, Z. et al. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proc. IEEE/CVF international conference on computer vision, 10012–10022 (2021).
Touvron, H., Cord, M. & Jégou, H. Deit iii: Revenge of the vit. In European conference on computer vision (ed. Touvron, H.) 516–533 (Springer, 2022).
Chen, J. et al. Transunet: Transformers make strong encoders for medical image segmentation. Preprint at arXiv:2102.04306 (2021).
Gu, A. & Dao, T. Mamba: Linear-time sequence modeling with selective state spaces. Preprint at arXiv:2312.00752 (2023).
Beck, M. et al. xlstm: Extended long short-term memory. Preprint at arXiv:2405.04517 (2024).
Zhu, L. et al. Vision mamba: Efficient visual representation learning with bidirectional state space model. Preprint at arXiv:2401.09417 (2024).
Berseth, M. Isic 2017-skin lesion analysis towards melanoma detection. Preprint at arXiv:1703.00523 (2017).
Codella, N. et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). Preprint at arXiv:1902.03368 (2019).
Jha, D. et al. Kvasir-seg: A segmented polyp dataset. In MultiMedia modeling: 26th international conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, proceedings, part II 26 (ed. Jha, D.) 451–462 (Springer, 2020).
Bernal, J. et al. Wm-dova maps for accurate polyp highlighting in colonoscopy: Validation vs saliency maps from physicians. Comput. Med. Imaging Graph. 43, 99–111 (2015).
Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. Preprint at arXiv:1804.03999 (2018).
Iqbal, A. & Sharif, M. Unet: A semi-supervised method for segmentation of breast tumor images using a u-shaped pyramid-dilated network. Expert Syst. Appl. 221, 119718 (2023).
Heinrich, M. P., Stille, M. & Buzug, T. M. Residual u-net convolutional neural network architecture for low-dose ct denoising. Curr. Direct. Biomed. Eng. 4, 297–300 (2018).
Vaswani, A. Attention is all you need. Advances in Neural Information Processing Systems (2017).
Cao, H. et al. Swin-unet: Unet-like pure transformer for medical image segmentation. In European conference on computer vision (ed. Cao, H.) 205–218 (Springer, 2022).
Wang, H., Cao, P., Wang, J. & Zaiane, O. R. Uctransnet: Rethinking the skip connections in u-net from a channel-wise perspective with transformer. Proc. AAAI Conf. Artif. Intell. 36, 2441–2449 (2022).
Gu, A. et al. Combining recurrent, convolutional, and continuous-time models with linear state space layers. Adv. Neural. Inf. Process. Syst. 34, 572–585 (2021).
Gu, A., Goel, K. & Ré, C. Efficiently modeling long sequences with structured state spaces. Preprint at arXiv:2111.00396 (2021).
Wang, Z., Zheng, J.-Q., Zhang, Y., Cui, G. & Li, L. Mamba-unet: Unet-like pure visual mamba for medical image segmentation. Preprint at arXiv:2402.05079 (2024).
Ruan, J. & Xiang, S. Vm-unet: Vision mamba unet for medical image segmentation. Preprint at arXiv:2402.02491 (2024).
Hochreiter, S. & Schmidhuber, J. Lstm can solve hard long time lag problems. Adv. Neural Inf. Process. Systems 9 (1996).
Al-Selwi, S. M., Hassan, M. F., Abdulkadir, S. J. & Muneer, A. Lstm inefficiency in long-term dependencies regression problems. J. Adv. Res. Appl. Sci. Eng. Technol. 30, 16–31 (2023).
Salehin, I. et al. Real-time medical image classification with ml framework and dedicated cnn-lstm architecture. J. Sensors 2023, 3717035 (2023).
Shahzadi, I., Tang, T. B., Meriadeau, F. & Quyyum, A. Cnn-lstm: Cascaded framework for brain tumour classification. In 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES) (ed. Shahzadi, I.) 633–637 (IEEE, 2018).
Chen, T. et al. xlstm-unet can be an effective 2d & 3d medical image segmentation backbone with vision-lstm (vil) better than its mamba counterpart. Preprint at arXiv:2407.01530 (2024).
Fan, D.-P. et al. Pranet: Parallel reverse attention network for polyp segmentation. In International conference on medical image computing and computer-assisted intervention (ed. Fan, D.-P.) 263–273 (Springer, 2020).
Loshchilov, I. Decoupled weight decay regularization. Preprint at arXiv:1711.05101 (2017).
Loshchilov, I. & Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. Preprint at arXiv:1608.03983 (2016).
Li, C. et al. U-kan makes strong backbone for medical image segmentation and generation. Preprint at arXiv:2406.02918 (2024).
Ruan, J., Xie, M., Gao, J., Liu, T. & Fu, Y. Ege-unet: an efficient group enhanced unet for skin lesion segmentation. In International conference on medical image computing and computer-assisted intervention (ed. Ruan, J.) 481–490 (Springer, 2023).
Funding
This work was supported by the Funds for Central - Guided Local Science & Technology Development (Grant No.202407AC110005) Key Technologies for the Construction of a Whole-process Intelligent Service System for Neuroendocrine Neoplasm
Author information
Authors and Affiliations
Contributions
Methodology,Software,Validation,X.Z; Founding acquisition,Resources,Supervision,G.L; Methodology,Writing-Review,H.L
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhong, X., Lu, G. & Li, H. Vision Mamba and xLSTM-UNet for medical image segmentation. Sci Rep 15, 8163 (2025). https://doi.org/10.1038/s41598-025-88967-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88967-5