Abstract
Oecomys (Rodentia, Sigmodontinae, Oryzomyini) is a taxonomically complex and cytogenetically diverse genus with a controversial intraspecific phylogenetic relationship. Karyotypic analyses, using whole chromosome probes from Hylaeamys megacephalus (HME, Sigmodontinae, Oryzomyini) in some taxonomic lineages of this genus have detected the rearrangements that shaped their karyotypes, in addition to revealing relevant insights into the taxonomic status of these taxa. Thus, to investigate the chromosomal evolution of the genus, we characterized the karyotype of Oecomys rutilus (ORU) with HME probes, establishing chromosomal homology maps with the karyotypes of other congeners. The chromosomal phylogeny obtained by Maximum Parsimony analysis recovered the genus Oecomys as monophyletic, with moderate bootstrap support (68%). This clade branches into two large groups, the first including O. rutilus followed by O. catherinae—Rio de Janeiro and O. catherinae—Pará; the other group includes O. auyantepui followed by O. paricola cytotype C and O. paricola cytotype A + cytotype B. We detected that these taxa underwent intensive reorganization of their karyotypes, the rearrangements producing this diversity were 15 pericentric inversions, 12 centric fusions, 11 fissions, 5 in tandem fusions, 8 translocations and the addition/deletion of constitutive heterochromatin on two autosomes and the X chromosomes. However, despite the high number of chromosomal rearrangements found, we identified some entirely conserved syntenic blocks shared among all species analyzed. From these data, we hypothesized a putative ancestral karyotype. We also detected exclusive characteristics for ORU, the syntenic blocks HME 1/20/4 (ORU 1), HME (16,17)/3 (ORU 2), HME 2/1 (ORU 4), HME 2/7 (ORU 5), HME 5/(9,10)/8 (ORU 3), HME 19/12 (ORU 9). We provide an overview of the chromosomal reorganization of the genus that points to a high chromosomal diversity and show that chromosomal rearrangements play an important role in the radiation of these species.
Similar content being viewed by others
Introduction
The genus Oecomys Thomas, 1906 (Cricetidae, Sigmodontinae, Oryzomyini) comprises 20 species of tree rats distributed throughout southern Central and South America1,2,3,4,5. The monophyly of the genus has been recovered through molecular and morphological analyses, and the existence of species complexes within some clades has been proposed; consequently, the real number of species remains inconclusive3,4,6,7,8,9,10,11,12,13,14,15.
Karyotypic information on chromosome number and morphology has been used for better taxonomic delineation at the intra and interspecific levels6,7,10,14,16. Oecomys presents a wide karyotypic variety, with diploid number (2n) ranging from 54 to 86 and autosomal fundamental number (FNa) from 54 to 1426,7,13,14,16,17,18,19,20,21. The species Oecomys rutilus (Reddish Oecomys), one of the lineages recovered as a valid and distinct taxon of Oecomys14, has 2n = 54 and FNa = 86 [this study], 9019, one of the smallest 2n for the genus, with chromosomal morphology marked by a large proportion of biarmed chromosomes.
The use of whole chromosome probes allows the comparison of karyotypes from different taxa, and thus provides details about chromosomal reorganization and identifies shared syntenies, which act as chromosomal signatures, as illustrated in species of the Sigmodontinae subfamily (for example22,23,24,25,26,27,28). Shared syntenies can also be used as markers in the construction of topologies based on a matrix of chromosomal characters26,28,29,30,31. Chromosomal changes, when associated with molecular phylogenetic trees, indicate the direction of the evolution of chromosomal rearrangements31,32,33.
Chromosomal painting analyses using whole chromosome probes from Hylaeamys megacephalus (HME; Oryzomyini)34 have been useful in clarifying taxonomic and phylogenetic problems, expanding knowledge of the biodiversity of the subfamily Sigmodontinae, in addition to providing information on the chromosomal symplesiomorphies, synapomorphies and autapomorphies of 25 taxa of the subfamily13,19,20,23,24,25,26,27,30,31,34. In this context, the HME probes34 used in Oecomys assisted in the taxonomic delineation of O. catherinae, O. paricola, and O. auyantepui, allowed the proposition of species complexes previously undetected by morphological and molecular analyses13,19,20. Furthermore, they assisted in identifying chromosomal signatures unique to the genus Oecomys (HME (13,22)/21 and HME 1 fissioned in three blocks), and in the species analyzed within this group, O. auyantepui (HME 26/20/18), O. catherinae (HME (9,10)/14/5, 23/19/11 and 26/11) and O. paricola (HME 4/19)13,20,21.
Given the chromosomal complexity and diversity in Oecomys, we aimed to investigate the chromosomal evolution of the genus and provide chromosomal characters useful for future taxonomic delineation. To achieve this, we characterized the karyotype of the species O. rutilus using HME probes34 and we established chromosomal homology maps with the karyotypes of other species of the genus that had already been mapped with the same probes. Furthermore, we constructed a phylogeny based on chromosomal syntenies aiming to reveal the direction of chromosomal events in the genus Oecomys. Other Sigmodontinae taxa previously mapped with the same set of probes were included in the phylogenetic analysis.
Results
Cytogenetic analysis
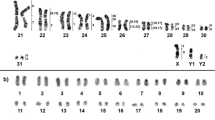
The Oecomys rutilus karyotype (ORU) has 2n = 54 and FNa = 86. The autosomal complement consists of 17 bi-armed chromosomal pairs ranging from large to small and nine medium to small acrocentric pairs. The X chromosome is a large subtelocentric and the Y is a medium submetacentric (Fig. 1 and Supplementary Fig. 1).
The hybridizations of HME probes to the Oecomys rutilus karyotype (2n = 54/FNa = 86, XY) revealed 41 homologous segments (Figs. 1 and 2 and Supplementary Table 1). Eleven probes showed conserved synteny with single signals in ORU, of which four (HME 18, 25, 24, 15) hybridized to whole chromosomes (ORU 15, 16, 21, 22, respectively), and seven (HME 4, 3, 8, 7, 21, 12, 26) to portions of chromosomes involved in chromosomal associations. Twelve HME probes (HME 1, 2, 5, 6, (9,10), 11, (13,22), 14, (16,17), 19, 20, 23) hybridized to more than one pair, and each fragment was identified with letters to facilitate understanding. Eleven ORU pairs (ORU 1, 2, 3, 4, 5, 6, 8, 9, 10, 12, 14) showed associations with HME probes (HME 1a/20a/4, HME (16,17)a/3, HME 5a/(9,10)/8, HME 2a/1b, HME 2b/7, HME (13,22)a/20b, HME 21/(13,22), HME 19a/12, HME 11a/26, HME 5b/14a, HME 19b/11b). Due to the poor quality of the hybridization, it was not possible to analyze the HME signals (13,22), but considering the hybridization gaps in the autosomes, we suggested their location in the ORU genome. The Supplementary Fig. 2 shows metaphases with hybridizations of all HME whole chromosome probes.
FISH results with HME whole chromosome probes in Oecomys rutilus. Probe identification is shown above the chromosomes and the numbers of the respective ORU chromosome pairs are shown below. When ORU chromosomes are presented in multiple images, syntenic associations are displayed with HME probes. *In Yellow: The hybridization gaps in the autosomes that led to the suggestions of the location of HME (13,22) in the ORU genome.
Phylogenetic analysis
From a total of 73,317 rearrangements, our analysis retrieved a single tree that showed the best score of 142 evolutionary steps. Bootstrap values ranged from 25 to 100% (Fig. 3).
Chromosomal phylogeny of 26 Sigmodontinae taxa analyzed using HME chromosomal probes. Tree based on the chromosomal character matrix. R. mastacalis was used as an outgroup. The numbers in each ramification represent the bootstrap value (only values above 50% are displayed). Abbreviations as in Supplementary Table 1. The characters are detailed in Oliveira da Silva et al.31.
The Sigmodontinae subfamily showed an initial branching with Rhipidomys mastacalis (RMA), followed by Rhipidomys emiliae (REM) (100%), both representatives of Thomasomyini; the next ramification (99%) recovered two groups with low branch support. The first group includes only Akodontini taxa, as follows: Thaptomys nigrita (TNI), Oxymycterus amazonicus (OAM), Necromys lasiurus (NLA), and Akodon diauarum (ADI) + Akodon montensis (AMO). Moderate to high bootstrap values supported the sister relationship between NLA and ADI + AMO (68%), and ADI + AMO (98%). The second group includes the Akodontini Blarinomys breviceps (BBR) as sister to a group composed of only Oryzomyini taxa, with 58% of branch support. Within this clade, the genus Cerradomys was recovered as sister to the genera Neacomys + Oecomys. All three genera appeared as monophyletic, with 97%, 88%, and 68% of bootstrap values, respectively. In the Cerradomys clade, C. scotii (CSC) was recovered as a sister to C. langguthi (CLA) and C. subflavus (CSU) + C. vivoi (CVI), with 72% of branch support, while CSU + CVI presented 69% of branch support. In the Neacomys clade, N. elieceri (NEL) appeared as sister to N. amoenus (NAM) + Neacomys sp. E (NSP-E), with 60% of branch support, while N. paracou (NPA) was recovered as sister to N. vossi and N. marajoara + N. xingu, the latter two ramifications with 68% of bootstrap values. The Oecomys clade also includes two subclades. The first one includes O. rutilus (ORU) as sister to O. catherinae from Rio de Janeiro (OCA-RJ) + O. catherinae from Pará (OCA-PA), with 64% of bootstrap value in the more inclusive node and 84% in the less inclusive node. The next subclade includes O. auyantepui as sister to O. paricola cytotype B (OPA-B) and O. paricola cytotypes A + C (OPA-A + OPA-C). Only the node that units the three cytotypes of O. paricola presented high support (95%). Considering the limited number of taxa and characters analyzed, species exhibited high levels of autapomorphy, resulting in low support values.
Discussion
Cytogenetic and distribution data for O. rutilus
The karyotype of O. rutilus (2n = 54/FNa = 86, 24 m + 8sm + 2st + 18a) obtained in this study (locality 1, Fig. 8) is similar to that described previously18 for O. rutilus (2n = 54/ FNa = 90, 24 m + 6sm + 8st + 14a) captured along the Negro River (Brazil, Amazonas, locality 2; Fig. 8). The FNa differences between these cytotypes are possibly due to pericentric inversions or centromeric repositioning in the smaller chromosomes, since the larger chromosomes have visibly conserved morphology. Although we could not perform a detailed comparison of the cytotypes, we do not exclude the hypothesis that other types of rearrangements may be involved in this differentiation. As was evidenced in O. paricola, whose cytotype B (2n = 70/FNa = 75) and cytotype C (2n = 70/FNa = 72) diverge only at FNa. In fact, comparative analysis by chromosome painting revealed that these cytotypes differed not only by pericentric inversions (or centromeric repositioning) but also by fusion/fission events and translocations20.
The karyomorphs described in the literature for Oecomys sp. 1 from Cuieiras River, Amazonas (2n = 54/NF = 84; 22 m + 8sm + 2st + 20a), Oecomys sp. 2 from Jatapu river, Amazonas (2n = 54/NF = 86; 24 m + 8sm + 2st + 18a) (localities 3 and 4, respectively; Fig. 8; Table 1)35, and O. cf. bicolor from Jari river, Pará (2n = 54/FNa = 82, 20 m + 8sm + 2st + 22a)35 may belong to O. rutilus because these cytotypes show high similarity to each other. Specifically, Oecomys sp. 2 shares the same karyotypic formula with our sample. Additionally, the occurrence localities of these karyomorphs are consistent with the distribution area of O. rutilus, which is restricted to the Guyana endemism center [see 2, 36]. These karyotypes are different from any others already described for species that occur in sympatry with O. rutilus, such as O. auyantepui (2n = 64, 65, 66, 72), O. rex (2n = 62/FNa = 80), and O. bicolor (2n = 80, 82, 86)2,6,19,21,35,36. However, genetic tools need to be used in conjunction with morphological analysis for taxonomic confirmation of individuals.
Chromosomal rearrangements in Oecomys
The comparative analysis by chromosome painting with HME probes among O. rutilus (ORU, 2n = 54/FNa = 86) and the species from the literature O. catherinae from Pará (OCA-PA, 2n = 62/FNa = 62); O. catherinae from Rio de Janeiro (OCA-RJ, 2n = 60/FNa = 62); O. paricola cytotype A (OPA-A, 2n = 72/FNa = 75); O. paricola cytotype B (OPA-B, 2n = 70/FNa = 75); O. paricola cytotype C (OPA-C, 2n = 70/FNa = 72) and O. auyantepui (OAU, 2n = 65/FNa = 84)13,19,20, revealed that their karyotypes are differentiated by a total of 53 chromosomal rearrangements: 15 pericentric inversions (PI), 12 centric fusions (FUS), 11 fissions (FIS), 5 in tandem fusions (tFUS), 8 translocations (Transl), addition/deletion of constitutive heterochromatin (CH) on 2 autosomes and the X chromosomes (Figs. 4, 5 and 6). Only one syntenic block was found without detectable rearrangements among these species, hybridized by the HME 24 probe (Figs. 4 and 5). Although they are phylogenetically related species14, extensive chromosomal rearrangements are responsible for the karyotypic diversification between them.
Idiograms of the haploid content of Oecomys catherinae from Pará (OCA-PA; 2n = 62/FNa = 62), O. catherinae from Rio de Janeiro (OCA-RJ; 2n = 60/FNa = 62)13, O. paricola cytotype A (OPA-A; 2n = 72/FNa = 75), O. paricola cytotype B (OPA-B; 2n = 70/FNa = 75), O. paricola cytotype C (OPA-C; 2n = 70/ FNa = 72)19, O. auyantepui (OAU; 2n = 65/FNa = 84)20, and O. rutilus (ORU; 2n = 54/FNa = 86), showing the organization of chromosomal syntenies identified by chromosome painting. Template adapted from28. The color code corresponds to the idiograms of the HME probe30, which is represented by the first chromosome (on the left) of each chromosome grouping.
Idiograms showing the chromosomal rearrangements involved in the karyotypic divergence between the species analyzed. Idiograms inside the box correspond to the karyotype of Hylaeamys megacephalus30.
Chromosomal signatures
Of the 13 chromosomal signatures proposed for the subfamily Sigmodontinae (HME 1a, 1b, 7/(9,10), 8, 1/12, 6/21, 11/(16,17), 5/(16,17), 20/(13,22), 15, 19/14/19, 24 and 26)24,25, ORU exhibits HME 15 and 24, in addition to the chromosomal signature 20/(13,22), which is found as a derived character, HME 20/(13,22) (ORU 6) and HME 1/20/4 (ORU 1). As mentioned earlier, all other Oecomys species share the character HME 24 with the subfamily, while the plesiomorphic character HME 20/(13,22) is entirely preserved only in OPA-B, in this same taxon HME 15 is found with an autapomorphy (15a and 15b) (Fig. 4). Some characters, absent in ORU, are observed in at least one of the Oecomys species (HME 1/12, 8, 14/19, 26).
Regarding the sex chromosomes, the long arm of the Y chromosome of ORU exhibited hybridization with the HME X probe. A possible explanation for this would be that this long arm is compsosed of the pseudoautosomal region and repetitive sequences shared with the X chromosome.
We confirmed the chromosomal synapomorphies proposed for the genus Oecomys, these being the HME 1 fragmented into three blocks, and the syntenic association HME (13,22)/2119 (see Fig. 4). The three fragments of HME 1 occur without any detectable rearrangement in the OCA and OPA group of species, where all hybridize to acrocentric chromosomes; in OAU, one of the fragments is involved in the HME 1/12 association (OAU 23); in ORU, we observe the associations HME 1/20/4 (ORU 1) and HME 2/1 (ORU 4), along with one of the fragments hybridizing in a metacentric chromosome (ORU 7). The syntenic block HME (13,22)/21 hybridizes acrocentric chromosomes in all species except ORU, which hybridizes a biarmed chromosome (ORU 8).
We also detected exclusive characteristics for ORU, the syntenic blocks HME 1/20/4 (ORU 1), HME (16,17)/3 (ORU 2), HME 2/1 (ORU 4), HME 2/7 (ORU 5), HME 5/(9,10)/8 (ORU 3), HME 19/12 (ORU 9), which are different from those described for OAU (26/20/18), OCA (HME (9,10)/14/5, 23/19/11) and OPA (HME 4/19); ORU shares the syntenic association HME 26/11 with OCA, previously described as exclusive to this species (OCA), although it is acrocentric in OCA and metacentric in ORU.
Phylogenetic relationship of the genus Oecomys from the perspective of chromosomal evolution
The chromosomal phylogeny obtained by Maximum Parsimony (MP) analysis recovered the genus Oecomys as monophyletic (Fig. 3). The species were grouped into two clades: (1) O. rutilus as sister to the O. catherinae karyotypic variants, and (2) O. auyantepui as sister to the O. paricola karyotypic variants (Fig. 6).
Comparing our phylogeny with that previously published31 it is possible to see that the inclusion of ORU data helped to define polytomies of the previous phylogeny. Thus, the relationships between OPA-A, OPA-B and OPA-C were defined, as well as those between NAM, NEL and NSP-E. There was also a change in the position of the species TNI. The other relationships remained the same. These changes are because the new data added probably changed the apomorphy/synapomorphy condition of some rearrangements. However, this arrangement found in the chromosome tree does not reflect the one recovered by the most thorough molecular topologies performed for Oecomys to date14. One of the explanations for this incongruity may be the number of species analyzed here; the other explanation would be the low support for both the molecular phylogeny and the chromosomal phylogeny, which may not represent the true relationship among the clade species. Furthermore, the association of molecular phylogeny14 with chromosomal characters turns the majority of synapomorphies into homoplasies or character reversions, therefore being less parsimonious than those that use the chromosomal approach.
Clade of Oecomys highlighted from the chromosomal topology of the subfamily Sigmodontinae (Fig. 3). Only the syntenies involved in rearrangements are represented in the idiograms (Node A). In branches B to M, chromosomal evolution is shown in O rutilus (ORU), O. catherinae from Pará (OCA-PA), O. catherinae from Rio de Janeiro (OCA-RJ), O. auyantepui (OAU), O. paricola cytotype A (OPA-A), O. paricola cytotype B (OPA-B) and O. paricola cytotype C (OPA-C). In each of the nodes, the numbers and types of rearrangements that occurred during taxon diversification are shown. FIS = fission; tFUS = in tandem fusion; FUS = centric fusion; Transl = translocation; PI = pericentric inversion; HC = constitutive heterochromatin.
According to Fig. 6, we identified 31 autosomal blocks involved in rearrangements (Node A): HME 3, 4, 6, 7, 20/(13,22), 8, (9,10), (9,10), 1, 1, (13,22)/21, 2, 2, 12, 19/14, 15, (16,17), (16,17), 18, 5, 11, 25, 26, 23, 23, 5, 11, 1, 19, 14, 5 (2 biarmed). Following the chromosomal topology groupings, we delineated the rearrangements that occurred during the branching of the species (Figs. 5 and 6). Thus, first a divergence occurred in two clades (nodes B and C). On the one hand, the phylogenetic branch (Node B) that groups ORU (Node D), OCA-RJ (Node H) and OCA-PA (Node I) is marked by one fission (HME 6a + 6b), two centric fusions (HME 11/26, 5/14) and one translocation (HME 14 + 19/11) that occurred before the divergence of these species; in ORU (Node D) there are five centric fusions ((16,17)/3, 2/1, 2/7, 19/12, 1/20b/4) [detailed description in Fig. 5], three translocations (resulting in HME 13,22/20a + 1/20b/4, (9,10)a + 5a + 5b/(9,10)b/8) [fragment “b” originates from the translocation of part of fragment “a”, it is not related in the fragments defined in Fig. 1: detailed description in Fig. 5], six pericentric inversions (HME 1, 21/(13,22), (9,10), (16,17), 18, 25); before the OCA-PA and OCA-RJ branch (Node E) there are three in tandem fusions (HME 2, (9,10)/14/5, 23/19/11), one pericentric inversion (HME 26/11) and one fission (HME 20/[13.22]a + [13.22]b); in OCA-RJ (Node H) there was one translocation (HME 2 + 2/3); in OCA-PA (Node I) we identified one fission (6b + 6c). On the other hand, in the phylogenetic branch (Node C) of the OAU (Node F), OPA-A (Node M), OPA-B (Node J), OPA-C (Node L) grouping, we detected two fissions (HME 4a + 4b; 3a + 3b) and one pericentric inversion (HME (16,17)); in this branch, the first taxon to diverge is OAU with two translocations (HME 6a + 6b/7, (9,10)b/(13,22)a + 26/20/18) [detailed description in Fig. 5]), two fissions (HME (9,10)a + (9,10)b/(13,22)a + (13,22)b [detailed description in Fig. 5]), five centric fusions (HME (9,10)/2, 1/12, 3/X, 26/20/18, (9,10)b/(13,22)a), four pericentric inversions (HME 6a, 4a, 23, 25). Before the diversification of the remaining taxa (Node G) we identified one in tandem fusion (HME 4/19), three pericentric inversions (HME (16,17), 5, 26 heteromorphic), addition of constitutive heterochromatin on two autosomes (HME (16 ,17), (16,17)); in OPA-B (Node J) one in tandem fusion (HME 11), two fissions (HME 15a + 15b; 25a + 25b); in the OPA-C and OPA-A grouping (Node K), there was one translocation (HME 20a/(13,22) + 20b/11), one fission (HME 20b/11a + 11b). In OPA-C (Node L) the homomorphic form of the homeologue HME 26 occurs, and in OPA-A (Node M) we identified one fission (HME 11a + 11b).
Putative ancestral karyotype
Based on the sharing of characters and using the other taxa of the Sigmodontinae subfamily as outgroup, we propose a putative ancestral karyotype (PAK) for the Oecomys species analyzed here. The PAK has 2n = 66/FNa = 66 (31 acro + 1 meta + X acro) (Fig. 7) and is composed of the previously mentioned autosomal blocks: HME 1a, 1b, 1c, 2a, 2b, 3, 4, 5a, 5b, 5c, 6, 7, 8, (9,10)a, (9,10)b, 11a, 11b, 12, (13,22)/21, 14, 15, (16,17)a, (16,17)b, 18, 19, 19/14, 20/(13,22), 23a, 23b, 25, 26, along with HME 24, which was conserved across all species (see Fig. 5).
Idiograms of the karyotypes of O. catherinae from Pará, O. catherinae from Rio de Janeiro, O. paricola cytotype A, O. paricola cytotype B, O. paricola cytotype C, O. auyantepui13,20,21, O. rutilus and the Putative Ancestral Karyotype (present study). “H” indicates the presence of constitutive heterochromatin. The color code corresponds to the HME probe, which is identified by the numbers next to each chromosome representative; model developed by Oliveira da Silva et al.30.
We highlight here that OCA-PA underwent 10 events of chromosomal rearrangements, involving 12 pairs of PAK chromosomes, to establish the current karyotype; OCA-RJ experienced 10 rearrangements in 13 pairs; OPA-A underwent 12 rearrangements in nine pairs; in OPA-B there were 13 rearrangements in 11 pairs; OPA-C experienced 12 rearrangements in 10 pairs; OAU presented 16 rearrangements in 16 pairs; lastly, ORU underwent 18 rearrangements involving 27 chromosomes from the proposed ancestor (Figs. 5, 6 and 7).
From the number of rearrangements and chromosomes involved, it is clear that the species exhibited different rates of chromosomal alterations. OPA and OCA are the taxa that share the most conserved chromosomal signals with PAK, while ORU, followed by OAU, underwent complex karyotype remodeling with intense rates of rearrangements (Figs. 5, 6 and 7). Pericentric inversions, together with centric fusions, were mainly responsible for chromosomal reorganization and the consequent increase in the number of chromosome arms in ORU and OAU. In ORU, centric fusions influenced the decrease in the diploid number, while in OAU, the proportion between fissions and centric fusions preserved the diploid number close to that of PAK. As the events occur in different syntenies, the increase in FNa in these two species occurred autonomously and could be a trend for the genus.
Despite carrying unique chromosomal characteristics, the cytotypes analyzed share significant similarities. We identified syntenic blocks that are conserved entirely across all analyzed species (HME 1a, 1b, 1c, 7, 8, (9,10), 12, 14, (16,17), (16,17), 18, 19, (13,22)/21, 23a, 23b, 24, 26) (Fig. 4), which are mainly involved in centric fusions and pericentric inversions (Fig. 6). The remaining chromosomes have varying numbers of signals among species, due to fissions and translocations in at least one species, and they are largely responsible for the diversity of 2n. This demonstrates that the evolutionary events that differentiate the karyotypes of these species, although independent, are not random.
Evolutionary trend for the genus Oecomys
The most thorough phylogenetic analyses available for Oecomys14 include the O. matogrossensis and O. rex species, along with the species groups O. paricola, O. rutilus, and O. auyantepui, as well as the species group O. catherinae. The PAK (2n = 66/FNa = 66) is in the range of 2n/FNa (2n = 54 to 72; FNa = 54 to 90) found in these species. We thus expand the PAK as existing prior to these species’ diversification.
Furthermore, we suggest that the conserved syntenic blocks (HME 1a, 1b, 1c, 7, 8, (9,10), 12, 14, (16,17), (16,17), 18, 19, (13,22)/21, 23a, 23b, 24, 26) present in OCA, OPA, OAU, ORU may also be present in the other taxa of the clade, remaining intact throughout the speciation of these species, shared as entire chromosomes, syntenic associations or pericentric inversions. This can be corroborated by the karyotypic structure found in O. matogrossensis (2n = 54/FNa = 54)14 and O. rex (2n = 62/FNa = 86)35, after an analysis of chromosomal morphology. We propose that in tandem fusions support the karyotypic organization of O. matogrossensis (2n = 54/FNa = 54), reducing both 2n and FNa; while the karyotypic diversity mechanism of O. rex (2n = 62/FNa = 86)36 could follow the pattern of rearrangements described here for O. auyantepui, a combination of pericentric inversions, centric fusions, fissions, and translocations, which would provide a balance between a minimum variation in 2n and an increase in FNa, in relation to PAK.
An ancestral karyotype with 2n = 60/FNa = 62 has been suggested for the Oecomys genus17. This suggestion was based on the wide geographic distribution of this cytotype. It is proposed that other karyotypes could have evolved from this ancestral karyotype through Robertsonian rearrangements and pericentric inversions17. However, here we demonstrate that chromosomal evolution in Oecomys is more complex than previously assumed, and the genome of this group is shaped by a combination of in tandem fusions, Robertsonian rearrangements, translocations, pericentric inversions and addition/deletion of constitutive heterochromatin. These rearrangements, in most cases, occur independently and involve different syntenies, providing an impressive karyotypic and genomic diversity of the genus.
Conclusion
Oecomys karyotypes exhibit significant variability in form, number, and chromosomal architecture, as demonstrated by comparative chromosome painting analysis. Different syntenic block combinations and the accumulation of various numbers and kinds of chromosomal rearrangements are the causes of this variety. The analysis indicates different rates of chromosomal changes between species, with only one chromosome entirely conserved. Many syntenic blocks were nevertheless retained despite this. The chromosomal phylogeny shows that O. rutilus shares chromosomal apomorphies with the karyotypic variants of O. catherinae, while O. auyantepui shares them with the karyotypic variants of O. paricola. Here, we show how important rearrangements were to Oecomys’ diversification; these were probably one of the factors that contributed to the genus’s radiation, since there is now evidence that chromosomal rearrangements play a role in speciation, as suggested by different theoretical models and supported by empirical microevolutionary studies37,38. Extending investigations to other species - especially those with a high diploid number - will be important to gain a better understanding of karyotypic evolution within the genus. It is worth noting that the genus Oecomys is currently subject to taxonomic revisions, and analysis of the phylogenetic relationships between its species, and studies of its geographic distribution. Chromosomal painting may be useful in these investigations because species-specific chromosomal signatures can serve as taxonomic markers and provide important information for species identification.
Methods
Sample
Our sample included 4 adult individuals of Oecomys rutilus (2 males and 2 females) collected in the municipality of Óbidos, Pará, Brazil (Fig. 8; Table 1). The taxonomic identification of the samples was verified using the identification key proposed by Carleton and Musser2. The skulls and skins were deposited in the zoological collection of the Zoology Museum of the Federal University of Pará (Vouchers: MUFPA 2024, MUFPA 2025, MUFPA 2028, MUFPA 2030).
The map (Fig. 8) was made in the software QGIS v. 3.10.7 (https://www.qgis.org/pt_BR/site/forusers/download.html). The database contains geographical data obtained from the DIVA-GIS platform39, available at https://www.diva-gis.org/gdata. Additional information is presented in Table 1.
Map with Amazonia areas of endemism and Marajo Island, collection points and cytogenetic data (2n/FNa) available in the literature and analyzed in the present study for O. rutilus. The numbers refer to the locations shown in Table 1.
Cytogenetics
Chromosomal preparations were obtained in vivo from bone marrow40. Metaphases were analyzed after G-banding41. Whole chromosome probes from Hylaeamys megacephalus (HME; 2n = 54/FN = 62, XX) were previously obtained by flow sorting at the Cambridge Resource Centre for Comparative Genomics, Department of Veterinary Medicine, University of Cambridge, UK34. Of the 24 probes produced, 21 corresponded to individual chromosome pairs and three corresponded to two pairs [HME (9,10), HME (13,22) and HME (16,17)]. The probes were labeled with biotin-16-dUTP ((Boehringer Mannheim), fluorescein isothiocyanate (FOTC)-12-dUTP (Amersham), or Cy3-dUTP by degenerate oligonucleotide primed PCR (DOP-PCR), as previously described34.
In situ hybridization of HME probes was performed as previously described34,42. Briefly, O. rutilus mitotic chromosome preparations were denatured in 70% formamide/2SSC at 65 °C for 50s. HME probes were denatured for 15 min at 70 °C. Hybridization was performed for 72 h at 37 °C. Post-hybridization washes included 2 × 50% formamide, 2 × 2SSC, 1 × 4SSC/Tween at 40 °C. Biotinylated probes were detected with avidin-Cy3 (red) or avidin-FITC (green) in single-color FISH experiments. The slides were counterstained with DAPI (4’,6-diamidino-2-phenylindole; blue). At least ten metaphases were analyzed in each sample by these techniques.
Image capture and analysis
Digital images of karyotypes with classical cytogenetics were obtained using an Olympus BX41 microscope with a CCD 1300QDS digital camera and analyzed using GenASIs software v. 7.2.7.34276 from ASI (Applied Spectral Imaging). FISH images were captured using AxioVision 3.0 software and a CCD camera (AxioCam) attached to a Zeiss-Axiophot 2 microscope or with NisElements software attached to a Nikon H550S microscope. For the correct assignment of hybridized chromosomes, we used inverted DAPI staining (G-band pattern) (Supplementary Fig. 3). The final images were edited using Adobe Photoshop CS6 software.
Comparative analysis
Chromosome painting data of the species O. catherinae from Pará (OCA-PA; 2n = 62/FNa = 62), O. catherinae from Rio de Janeiro (OCA-RJ; 2n = 60/FNa = 62), O. paricola cytotype A (OPA-A; 2n = 72/FNa = 75), O. paricola cytotype B (OPA-B; 2n = 70/FNa = 75), O. paricola cytotype C (OPA-C; 2n = 70/FNa = 72), and O. auyantepui (OUA; 2n = 65/FNa = 84) were recovered from the literature for comparison purposes with O. rutilus [see: 13, 21, 22]. The detected FISH signals are described in Supplementary Table 1.
Phylogenetic analysis
Chromosomal morphology, number and syntenic blocks based on HME probes hybridized to metaphases of several Sigmodontinae taxa obtained herein and from previous chromosomal painting studies13,21,23,24,25,26,27,30,31,34 (Supplementary Table 1) were converted into a data matrix of 70 non-additive (unordered) multi-state characters (Supplementary Table 2) on Mesquite program version 3.7043, following previous criteria31. The matrix contains nine genera from three tribes, as follows: Akodon montensis (AMO), A. diauarum (ADI), Blarinomys breviceps (BBR), Necromys lasiurus (NLA), Oxymycterus amazonicus (OAM), and Thaptomys nigrita (TNI) from tribe Akodontini; Cerradomys langguthi (CLA), C. scotii (CSC), C. subflavus (CSU), C. vivoi (CVI), Hylaeamys megacephalus (HME), Neacomys amoenus (NAM), N. elieceri (NEL), N. marajoara (NMA), N. paracou (NPA), Neacomys sp. E (NSP-E), N. vossi (NVO), N. xingu (NXI), Oecomys auyantepui (OAU), O. catherinae from Pará (OCA-PA), O. catherinae from Rio de Janeiro (OCA-RJ), O. paricola cytotype A (OPA-A), O. paricola cytotype B (OPA-B), O. paricola cytotype C (OPA-C), and O. rutilus (ORU) from tribe Oryzomyini; Rhipidomys mastacalis (RMA) and R. emiliae (REM) from tribe Thomasomyini.
A Maximum Parsimony (MP) phylogenetic analysis was performed using T.N.T. (Tree analysis using New Technology) program version 1.644. Branch support values were calculated with 1000 bootstrap replicates. The exhaustive search was made using Tree Bisection Reconnection (TBR). Rhipidomys mastacalis (RMA) was used as outgroup. The tree was displayed and edited in the Figtree program version 1.4.2 http://tree.bio.ed.ac.uk/software/figtree/). Gaps were treated as “missing data.”
Data availability
Data is provided within the manuscript or supplementary information files.
Change history
07 March 2025
The original online version of this Article was revised: In the original version of this Article the ORCID ID for Patricia Caroline Mary O’Brien was incorrect. The ORCID ID for Patricia Caroline Mary O’Brien has been removed.
References
Musser, G. G. & Carleton, M. D. Superfamily Muroidea. In Mammal Species of the World: A Taxonomic and Geographic Reference (eds Wilson, D. E. & Reeder, D. M.) 3 edn., 894–1531 (John Hopkins University, 2005).
Carleton, M. D. & Musser, G. G. Genus Oecomys Thomas, 1906. In Mammals of South America: Rodents (eds. Patton, J. L. et al.) 2 edn., 393–417 (The University of Chicago Press, 2015).
Rocha, R. G. et al. Cryptic diversity in the Oecomys roberti complex: revalidation of Oecomys tapajinus (Rodentia, Cricetidae). J. Mammal. 99 (1), 174–186. https://doi.org/10.1093/jmammal/gyx149 (2017).
Saldanha, J. & Rossi, R. V. Integrative analysis supports a new species of the Oecomys catherinae complex (Rodentia, Cricetidae) from Amazonia. J. Mammal. 102 (1), 69–89. https://doi.org/10.1093/jmammal/gyaa145 (2021).
Saldanha, S. et al. Unveiling hidden diversity of Oecomys (Rodentia: Cricetidae) from Brazilian central Amazonia: description of a new species and new lineages. Syst. Biodivers. 21 (1), 37. https://doi.org/10.1080/14772000.2023.2259037 (2023).
Patton, J. L., Silva, M. N. & Malcolm, J. R. Mammals of the Rio Juruá and the evolutionary and ecological diversification of Amazonia. Bull. Am. Mus. Nat. Hist. 244, 1–306 (2000).
Andrade, A. F. B. & Bonvicino, C. R. A new karyological variant of Oecomys (Rodentia: Sigmodontinae) and its phylogenetic relationship based on molecular data. Genome 46 (2), 195–203. https://doi.org/10.1139/g02-123 (2003).
Weksler, M. Phylogeny of neotropical Oryzomyini rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Mol. Phylogenet Evol. 29 (2), 331–349. https://doi.org/10.1016/s1055-7903(03)00132-5 (2003).
Weksler, M. Phylogenetic relationships of Oryzomyini rodents (Muroidea, Sigmodontinae): separate and combined analyses of morphological and molecular data. Bull. Am. Mus. Nat. Hist. 296, 1–149 (2006).
Rosa, C. C. et al. Genetic and morphological variability in south American rodent Oecomys (Sigmodontinae, Rodentia): evidence for a complex of species. J. Genet. 91, 265–277. https://doi.org/10.1007/s12041-012-0182-2 (2012).
Rocha, R. G., Fonseca, C., Zhou, Z., Leite, Y. L. R. & Costa, L. P. Taxonomic and conservation status of the elusive Oecomys cleberi (Rodentia, Sigmodontinae) from central Brazil. Mamm. Biol. 77, 414–419. https://doi.org/10.1016/j.mambio.2012.02.004 (2012).
Pardiñas, U. F. J., Teta, P., Salazar-Bravo, J., Myers, P. & Galliari, C. A. A new species of arboreal rat, genus Oecomys (Rodentia, Cricetidae) from Chaco. J. Mammal. 97, 1177–1196. https://doi.org/10.1093/jmammal/gyw070 (2016).
Malcher, S. M. et al. Oecomys catherinae (Sigmodontinae, Cricetievidenceidence for chromosomal speciation? PLoS ONE. 12 (7), e0181434. https://doi.org/10.1371/journal.pone.0181434 (2017).
Suárez-Villota, E. Y., Carmignotto, A. P., Brandão, M. V., Percequillo, A. R. & Silva, M. J. J. Systematics of the genus Oecomys (Sigmodontinae: Oryzomyini): molecular phylogenetic, cytogenetic and morphological approaches reveal cryptic species. J. Mammal. 97 (4), 1177–1196 (2017).
Saldanha, J. et al. Genetic diversity of Oecomys (Rodentia, Sigmodontinae) from the Tapajós River basin and the role of rivers as barriers for the genus in the region. Mamm. Biol. 97, 41–49. https://doi.org/10.1016/j.mambio.2019.04.009 (2019).
Andrades-Miranda, J., Oliveira, L. F. B., Zanchin, N. I. T. & Mattevi, M. S. Chromosomal description of the rodent genera Oecomys and Nectomys from Brazil. Acta Theriol. 46, 269–278 (2001).
Langguth, A., Maia, V. & Mattevi, M. Karyology of large size Brazilian species of the genus Oecomys Thomas, 1906 (Rodentia, Muridae, Sigmodontinae). Arq. Mus. Nac. 63, 183–190 (2005).
Asfora, P. H., Palma, A. R. T., Astúa, D. & Geise, L. Distribution of Oecomys catherinae Thomas, 1909 (Rodentia: Cricetidae) in northeastern Brazil with karyotypical and morphometrical notes. Biota. Neotrop. 11, 415–424. https://doi.org/10.1590/S1676-06032011000200039 (2011).
Gomes Júnior, R. G. et al. Intense genomic reorganization in the genus Oecomys (Rodentia, Sigmodontinae): comparison between DNA barcoding and mapping of repetitive elements in three species of the Brazilian Amazon. Comp. Cytogen. 10 (3), 401–426. https://doi.org/10.3897/CompCytogen.v10i3.8306 (2016).
Oliveira da Silva, W. et al. Karyotypic divergence reveals that diversity in the Oecomys paricola complex (Rodentia, Sigmodontinae) from eastern Amazonia is higher than previously thought. PLoS ONE. 15 (10), e0241495. https://doi.org/10.1371/journal.pone.0241495 (2020a).
Oliveira da Silva, W. et al. The emergence of a new sex-system (XX/XY1Y2) suggests a species complex in the monotypic rodent Oecomys auyantepui (Rodentia, Sigmodontinae). Sci. Rep. 12, 8690. https://doi.org/10.1038/s41598-022-12706-3 (2022).
Ventura, K., O’Brien, P. C. M., Yonenaga-Yassuda, Y. & Ferguson-Smith, M. A. Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: the evolution of the lowest diploid number in rodents. Chromosome Res. 17, 1063–1078. https://doi.org/10.1007/s10577-009-9083-5 (2009).
Suárez, P. et al. Clues on syntenic relationship among some species of Oryzomyini and Akodontini tribes (Rodentia: Sigmodontinae). PLoS ONE. 10 (12), e0143482. https://doi.org/10.1371/journal.pone.0143482 (2015).
Pereira, A. L. et al. Extensive chromosomal reorganization among species of New World muroid rodents (Cricetidae, Sigmodontinae): searching for phylogenetic ancestral traits. PLoS ONE. 11 (1), e0146179. https://doi.org/10.1371/journal.pone.0146179 (2016).
Oliveira da Silva, W. et al. Chromosomal diversity and molecular divergence among three undescribed species of Neacomys (Rodentia, Sigmodontinae) separated by amazonian rivers. PLoS ONE. 12 (8), e0182218. https://doi.org/10.1371/journal.pone.0182218 (2017).
Oliveira da Silva, W. et al. Chromosomal signatures corroborate the phylogenetic relationships within Akodontini (Rodentia, Sigmodontinae). Int. J. Mol. Sci. 21 (7), 2415. https://doi.org/10.3390/ijms21072415 (2020b).
Paixão, V. D. S. et al. Comparative genomic mapping reveals mechanisms of chromosome diversification in Rhipidomys species (Rodentia, Thomasomyini) and syntenic relationship between species of Sigmodontinae. PLoS ONE. 16 (10), e0258474. https://doi.org/10.1371/journal.pone.0258474 (2021).
do Nascimento M. C., Percequillo, C., Ferguson-Smith, A. R., Yonenaga-Yassuda, M. A. & Ventura, Y. Chromosomal evolution of tribe Oryzomyini (Rodentia: Cricetidae: Sigmodontinae). Mamm. Biol. 102 (2), 441–464. https://doi.org/10.1007/s42991-022-00244-4 (2022).
Hass, I., Sbalqueiro, I. J. & Muller, S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from Southern Brazil established by Zoo-FISH using Mus musculus (Muridae) painting probes. Chromosome Res. 16, 75–88. https://doi.org/10.1007/s10577-007-1211-5 (2008).
Oliveira da Silva, W. et al. Chromosomal phylogeny and comparative chromosome painting among Neacomys species (Rodentia, Sigmodontinae) from eastern Amazônia. BMC Evol. Biol. 19, 184. https://doi.org/10.1186/s12862-019-1515-z (2019).
Oliveira da Silva, W. et al. Chromosomal rearrangements played an important role in the speciation of rice rats of genus Cerradomys (Rodentia, Sigmodontinae, Oryzomyini). Sci. Rep. 14, 545. https://doi.org/10.1038/s41598-023-50861-3 (2024).
Di-Nizo, C. B. et al. Comparative chromosome painting in six species of Oligoryzomys (Rodentia, Sigmodontinae) and the karyotype evolution of the genus. PLoS ONE. 10 (2), e0117579. https://doi.org/10.1371/journal.pone.0117579 (2015).
Di-Nizo, C. B., Ferguson-Smith, M. A. & Silva, M. J. J. Extensive genomic reshuffling involved in the karyotype evolution of genus Cerradomys (Rodentia: Sigmodontinae: Oryzomyini). Genet. Mol. Biol. https://doi.org/10.1590/1678-4685-GMB-2020-0149 (2020).
Nagamachi, C. Y. et al. FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotypes. Chromosome Res. 21 (2), 107–119. https://doi.org/10.1007/s10577-013-9341-4 (2013).
Lira, T. Citogenética clássica e molecular de alguns representantes da tribo Oryzomyini (Rodentia, Cricetidae) da Amazônia Central. M.Sc. Thesis, Instituto Nacional de Pesquisas da Amazônia, Brazil (2012).
Voss, R. S., Lunde, D. P. & Simmons, N. B. The mammals of Paracou, French Guiana: a Neotropical lowland rainforest fauna part 2. Nonvolant species. Bull Am Mus Nat Hist 263, 3–236 (2001).
Berdan, E. L. et al. Structural variants and speciation: multiple processes at play. Cold Spring Harb Perspect. Biol. https://doi.org/10.1101/cshperspect.a041446
Lucek, K. et al. The impact of chromosomal rearrangements in speciation: from micro- to macroevolution. Cold Spring Harb Perspect. Biol. https://doi.org/10.1101/cshperspect.a041447
Hijmans, R. J. et al. DIVA-GIS. Vsn. 5.0. A Geographic Information System for the Analysis of Species Distribution Data. http://www.diva-gis.org (2004).
Ford, C. E. & Hamerton, J. L. A colchicine hypotonic citrate squash sequence for mammalian chromosome. Stain Technol. 31 (6), 247–251. https://doi.org/10.3109/10520295609113814 (1956).
Sumner, A. T., Evans, H. J. & Buckland, R. A. New technique for distinguishing between human chromosomes. Nat. (Lond.) New. Biol. 31, 282. https://doi.org/10.1038/newbio232031a0 (1971).
Yang, F., Carter, N. P., Shi, L. & Ferguson-Smith, M. A. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103, 642–652. https://doi.org/10.1007/BF00357691 (1995).
Maddison, W. P. & Maddison, D. R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.40. http://mesquiteproject.org (2018).
Goloboff, P. A., Farris, J. S. & Nixon, K. C. T.N.T., a free program for phylogenetic analysis. Cladistics 24, 774–786 (2008).
Sikes, R. S. & Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of mammalogists for the use of wild mammals in research. J. Mammal. 97 (3), 663–688. https://doi.org/10.1093/jmammal/gyw078 (2016).
Acknowledgements
The authors are grateful to MSc. Jorge Rissino, to MSc. Shirley Nascimento and Maria da Conceição for assistance in laboratory work. The authors thank Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) and Secretaria de Estado de Meio Ambiente do Para´ (SEMA-PA) for the authorization of sample collections. This study is part of the Thesis of VSP in Genetics and molecular Biology, under a CAPES Scholarship. CYN (305880/2017-9, 307170/2021-7) and JCP (305876/2017-1, 307154/2021-1) are grateful to CNPq for Productivity Grants.
Funding
This research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Fundação Amazônia Paraense de Amparo à Pesquisa (FAPESPA) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) on projects coordinated by CY Nagamachi (Edital BIONORTE-CNPq, Proc 552032/2010-7; Edital BIONORTE-FAPESPA, ICAAF 007/2011; Edital Pró-Amazônia Proc 047/2012; Edital Jovens Doutores CNPQ-FAPESPA, termo do Outorga 006/2023); the FAPESPA (Edital Vale—Proc 2010/110447) and Banco Nacional de Desenvolvimento Econômico e Social—BNDES (Operação 2.318.697.0001), Edital Jovens Doutores CNPQ-FAPESPA, termo do Outorga 013/2023) on a project coordinated by JCP; CNPq for Productivity Grants to CYN (307170/2021-7) and to JCP (307154/2021-1); Edital Jovens Doutores CNPq-FAPESPA, termo de Outorga 013/2023 for Scholarship to WOS. The article processing charge was paid by PROPESP-UFPA (Edital 06/2021—PAPQ/PROPESP).
Author information
Authors and Affiliations
Contributions
VSP: Conceptualization; Data Curation; Formal analysis; Investigation; Methodology; Visualization; Writing original draft; Writing review and editing. SMM: Data Curation; Methodology; Visualization; Writing review and editing. WOS: Made the map. Visualization; Writing review and editing. MAF-S: Investigation; Methodology; Resources; Visualization; Writing review and editing. PCMO’B: Investigation; Methodology; Visualization; Writing review and editing. RVR: Contributed with samples; Investigation; Methodology; Visualization; Writing review and editing. JCP: Data Curation; Formal analysis; Funding acquisition; Resources; Visualization; Writing review and editing. CYN: Project administration; Data Curation; Formal analysis; Funding acquisition; Resources; Supervision; Visualization; Writing review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The specimens were collected according to the guidelines recommended by the American Mammal Society45. The collection was carried out under JCP´s license number 13248, issued by the “Instituto Chico Mendes de Conservação da Biodiversidade”. The Cytogenetics Laboratory at CEABIO, UFPA, is authorized by the Ministry of the Environment for the transportation of samples (number 19/2003) and the use of samples for research (number 52/2003). This research was approved by the Ethics Committee of the Federal University of Pará (Permission 68/2015). The animals were euthanized using intraperitoneal injection of barbiturates (120 mg/kg pentobarbital) after applying local anesthesia (topical application of lidocaine). This study is performed in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
dos Santos Paixão, V., Malcher, S.M., Oliveira da Silva, W. et al. Chromosomal rearrangements drive diversity in arboreal rodents of the genus Oecomys. Sci Rep 15, 6111 (2025). https://doi.org/10.1038/s41598-025-89517-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-89517-9