Abstract
Accurately segmenting the aneurysm area from CTA data can reconstruct the three-dimensional morphology of the aneurysm, effectively evaluating the type, size, and risk of rupture of the aneurysm. However, accurate separation of the aneurysm is limited by the accuracy of image segmentation algorithms. Currently, the segmentation methods for intracranial aneurysms using CTA big data and deep learning lack universality. When faced with a new hospital acquired imaging modality, it is usually necessary to redesign and train the segmentation network. In response to this issue, this article proposes a more universal segmentation model and develops the GLIA Net algorithm (MGLIA Net model) based on MoblieNet, which can perform adaptive target segmentation on aneurysm images collected under different conditions. To verify the effectiveness of the algorithm in intracranial aneurysm segmentation, performance tests were conducted on an open-source dataset. The results showed that the proposed algorithm achieved segmentation accuracy of 55.9% and 73.1% on two datasets, respectively, significantly better than the original GLIA-Net algorithm.
Similar content being viewed by others
Introduction
An Intracranial aneurysm (IA), also known as a brain aneurysm1,2,3, is an abnormal weak spot in the wall of a blood vessel that causes an outward bulge, similar to a bubble. A bulging aneurysm can exert pressure on nerves or surrounding brain tissue and may cause leakage or rupture, spilling blood into the surrounding tissue (known as bleeding). An aneurysm can affect any blood vessel in the body, but only the blood vessels in the head can cause hemorrhagic stroke, which is a serious disease that can lead to brain damage and death after rupture. The commonly used neuroimaging techniques for intracranial aneurysms include digital subtraction angiography (DSA), magnetic resonance angiography (MRA), computed tomography angiography (CTA), and transcranial Doppler ultrasound examination4,5,6. Among them, CTA has high resolution, can clearly display vascular details in various parts, and is non-invasive and easy to operate. It is often used as a first-line diagnostic method for suspected intracranial aneurysm patients7,8.
Accurately segmenting the aneurysm area from CTA data can reconstruct the three-dimensional morphology of the aneurysm, effectively assessing its type, size, and risk of rupture. However, the accurate segmentation of aneurysms is limited by the accuracy of image segmentation algorithms.At present, researchers have developed various aneurysm segmentation methods based on big data and deep learning, and U-Net neural network is one of the widely used networks in the field of medical image segmentation. The concept of fully convolutional networks was initially proposed and introduced by Ronneberger et al.9. Stember et al.10 used the U-Net al.gorithm to detect and segment intracranial aneurysms in MIP images of MRA. Compared with the results marked by radiologists, its sensitivity was 98.8% (85/86), with differences in size and area of 2.01 mm (30%) and 8.21mm2 (27%), respectively. Segmentation of intracranial aneurysms can not only quickly determine intracranial aneurysms, but also further study the morphology of intracranial aneurysms.
One of the characteristics of deep learning networks is that the deeper the network layers, the stronger their learning ability. But the problem is also very obvious, as the depth of the network increases, there is not only a gradient vanishing problem, but also a more urgent degradation problem. This means that as the depth increases, the segmentation accuracy will rapidly decrease after reaching saturation. He et al.11 developed Convolutional Residual Networks (CRNs), which enable the network to achieve accuracy from deeper learning by providing a residual graph to the stacked layers every few layers.
Yuki et al.12 used ResNet-18 network to detect intracranial aneurysms in MIP images in MRA in their case series. Their algorithm detected five aneurysms smaller than 2 mm that had been missed by two radiologists, which could be used by younger doctors to diagnose intracranial aneurysms. Ueda et al.13 proposed using an 18 layer CNN residual network neural architecture for detecting intracranial aneurysms in MRA images from different institutions and multiple branded devices. The sensitivity of the algorithm to intracranial aneurysms in internal and external datasets was 91% and 93%, respectively. The algorithm has good compatibility to different mechanisms and different models. However, for intracranial aneurysms with a diameter greater than 5 mm, due to uneven internal signals in MRA images, their sensitivity was poor and segmentation of intracranial aneurysms has not been achieved. Park et al.14 developed a 3D CNN named HeadXNet, whose encoder was adapted from the 50-layer SE-ResNeXt network to detect intracranial aneurysms from CTA images. 328 positive CT angiography cases and 490 negative CT angiography cases were studied. Their model performed well in terms of sensitivity, accuracy, and consistency between doctors. Bo et al.15 proposed a 3D U-net called GLIA Net, which takes the entire original CTA scan (global image) as input and does not require preprocessing. The detection and segmentation of intracranial aneurysms from 1338 CTA images yielded an overall sensitivity of 90% and a false positive rate of 4 per case. GLIA-Net outperforms HeadXNet and U-Net models in segmentation performance.
To further improve network performance, this paper introduces the depthwise separable convolution from MobileNet for the three-dimensional structural features of CTA images16. This convolution decomposes the standard convolution into two steps: depthwise convolution focuses on extracting spatial features, while pointwise convolution focuses on fusing channel features. This article proposes a MGLIA-Net al.gorithm based on MoblieNet, which can better capture structural information in three-dimensional space when processing CTA images, thereby improving segmentation accuracy. Specifically, since separable convolution significantly reduces the amount of computation, deeper networks can be constructed without worrying about the risk of overfitting, and deeper networks can usually capture richer details, thereby improving the detection accuracy of intracranial aneurysms.
Methods
MGLIA-Net model
Based on the research of Ronneberger et al.17, organ and lesion region segmentation is one of the important contents in medical image analysis. Traditional methods usually require the establishment of a separate segmentation model for each organ and each modality. When faced with a new imaging modality collected by hospitals, it is usually necessary to retrain and design segmentation networks, which lacks universality. Therefore, a more universal segmentation model is designed in this paper, and one model is used to achieve adaptive target segmentation of aneurysm images collected under different patient demographics (age, sex) and aneurysm characteristics (size, location, morphology). For the target segmentation of intracranial aneurysms, which has a three-dimensional structure, it is necessary to consider the spatial correlation from different domain images and model cross channel correlation convolution. Therefore, the separable convolution idea is very suitable for the segmentation of such targets. There are significant differences in the number and morphology of intracranial aneurysms among different patients, as well as in the images captured by different hospitals. Separable convolution exhibits better generalization ability due to its parameter minimization, which means it can achieve better results when processing different CTA data. In addition, this paper introduces a certain attention mechanism, namely the global positioning module mentioned in reference18.
MobileNet V2 module
In the deep convolution stage, MobileNet applies convolution only to individual input channels, creating a lightweight network structure. Depthwise separable convolution further processes the output from depthwise convolution using pointwise convolution, yielding the final feature map. The proposed method significantly reduces computational complexity and parameter count compared to traditional convolution, improving training and inference speed while maintaining high accuracy, thus facilitating easier deployment.
Figure 1 depicts the conventional convolution process, where a convolution kernel of a defined shape leads to a significant number of parameters and computational workload for each application. In contrast, depthwise separable convolution breaks the process into two stages: depthwise convolution (DW) and pointwise convolution (PW), as shown in Figs. 2 and 3. Depthwise convolution processes each channel independently, resulting in a feature map with the same channel count as the original. Since it cannot utilize information across channels simultaneously, pointwise convolution combines features across channels. Overall, depthwise separable convolution requires significantly less computation than traditional convolution, enhancing processing speed.
Schematic diagram of residual block and inverted residual block16.
Figure 4 illustrates residual and inverted residual blocks. In Fig. 4(a), the residual block has more channels in the residual connection than in the middle convolution, employing a dimensionality reduction convolution, a feature extraction convolution, and a final dimensionality increase convolution, forming an hourglass shape. In contrast, Fig. 4(b) shows the inverted residual block, which first increases dimensionality, extracts features through channel-wise convolution, and then reduces dimensionality. This inversion creates a structure that is broader in the middle. Inverted blocks maintain efficiency while allowing for deeper network architectures.
The complete MobileNet V2 module is presented in Fig. 5. It reduces input channels using inverted residual blocks with a bottleneck layer followed by depthwise convolution for feature extraction and a final convolution to increase output channels. MobileNet V2 features linear bottlenecks, substituting nonlinear activation functions with linear ones, thereby reducing computational requirements and enhancing efficiency.
GLIA-Net framework
GLIA-Net (Global Localization-based Intracranial Aneurysm Network)15 is a neural network that integrates global localization with local segmentation for detecting intracranial aneurysms. It comprises two main components: the global localization network, which estimates aneurysm probabilities from the entire image, and the local segmentation network, which analyzes sub-images for precise aneurysm detection.
As shown in Fig. 6, the global feature generator processes the complete input image through five encoding blocks, producing feature maps with output channels of 8, 16, 32, 64, and 128, with downsampling in some layers. The global feature map is reshaped using ROI pooling to align with local image inputs, guiding the local segmentation network through skip connections.
The local segmentation network is structured as a U-Net with four encoding blocks (output channels: 16, 32, 64, 128). After downsampling, the outputs from the first three blocks are combined with the global features. The decoder consists of three blocks (output channels: 64, 32, 16) that restore the feature map to the original image size using transposed convolutions and residual blocks, culminating in a final convolution layer that produces the local segmentation map.
The loss function of the GLIA-Net model is divided into global loss and local loss functions, and the calculation formula is shown in (1):
Among them, \({\omega _{Global}}\) represents global weight,\({\omega _{Local}}\) represents local weight,\({L_{Global}}\) represents global loss, and \({L_{Local}}\) represents local loss.
The global loss calculation formula is shown in (2) :
Among them, \({z}\) represents the label, \(\widehat {z}\) represents the prediction result of the model.
The local loss calculation formulas are shown in (3), (4), and (5):
Among them, \({y}\) represents the label, \(\widehat {y}\) represents the model’s prediction results, \({\gamma _{Dice}}\)and\({\gamma _{Cross}}\)represent the nonlinearity of the control loss function, \(\varepsilon\) represents the smoothing factor, \({\omega _P}\) represents the pyramid weight.
MGLIA-Net framework
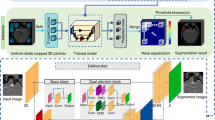
In this paper, an end-to-end convolutional neural network model with high accuracy, easy deployment and fast inference was developed by combining the characteristics of GLIA-Net and Mobile V2, and the specific distribution of intracranial aneurysms was predicted through the processing of three-dimensional brain CTA images, as shown in Fig. 7, which is the schematic diagram of MGLIA network.
This network is a 4-layer U-Net network model, the input is the local slice of CTA 3D image, and the output is corresponding to the prediction of intracranial aneurysm distribution. The network downsamples through maximum pooling, with encoder output channels of 8, 16, 32 and 64, respectively. On the basis of the traditional U-Net, the convolution layer is replaced by the separable convolution of Mobile V2. This convolution method first performs pointwise convolution on the input feature graph, increasing its dimensionality to twice the number of channels. Then, group them by the number of channels and perform depthwise convolution on them. Finally, the pointwise convolution reduces the number of feature graphs channels to the specified number of output channels. On this basis, the inverted residual mechanism is added to sum the input feature graph and the convolution feature graph to prevent gradient explosion during training. The decoder part is upsampled by transposing convolution, and the decoder output channels are 32, 16, and 8, respectively, corresponding to the encoder. The convolution layer in the decoder is still the Moblie V2 module with separable convolution plus inverted residuals. In the skip connection section, this paper introduces the global localization module in GLIA-Net, which preprocesses the global brain CTA image and provides a preliminary probability distribution graph of intracranial aneurysms. It is then multiplied point-by-point with the feature graph in the form of an attention mechanism and concatenated to the output of the decoder to guide U-Net to perform more refined object detection segmentation.
Data set information
IAs are difficult to identify due to their diversity in location, morphology, and size. This paper validates the effectiveness of the proposed algorithm using the dataset from reference15, which employs an open-source large-scale dataset containing data from different hospitals, patients, and lesions for network training and evaluation. The training and testing dataset consists of a total of 1338 CTA images, including 1489 intracranial aneurysm. These CTA images all contain the patient’s head area, some of which may also include the neck or heart area. The data included unruptured aneurysms and ruptured aneurysms with subarachnoid or parenchymal bleeding. During the network training phase, the data set was divided into 1186 training samples and 152 test samples. Internal test set with 50 negative cases (no IAs). Due to the small number of IAs in the brain, the training set al.ready had a serious data imbalance problem, so negative cases were not included in the training set. We verified that the distribution of all positive CTA images in the internal training and test sets was roughly the same across institutions, ages, and genders. The training set is used for model training, and the test set is only used to evaluate the model performance, ensuring that the model does not come into contact with the test set images during the training process.
To evaluate the generalization ability of the network model, the open-source data set also includes an external test set, divided into two parts, A and B. Test set A contains 71 cases (24 of which were negative), and test set B contains 67 cases (22 of which were negative). Detailed data set statistics are shown in Table 1.
Results
In order to verify the effectiveness of the proposed method, this paper compared it with other three methods, namely U-Net, HeadXNet and GLIA-Net. Firstly, compared with U-Net, this network model has been widely used in many segmentation tasks in the past few years and has made significant improvements compared to traditional models. Then, this model was compared with the HeadXNet network model. This model can be seen as adding a feature extraction network SE ResNiXt and a cordless spatial pyramid pooling module on the basis of U-Net. The reason for choosing this network for comparison is that it has shown great potential in segmentation tasks and its ability has also been tested on clinical data.Finally, this model was compared with the GLIA-Net network model, which made multiple improvements on the traditional U-Net structure, including the use of deeper network layers, stronger feature extraction capabilities, and more efficient fusion strategies.These improvements make GLIA Net perform better in handling complex medical image segmentation tasks. By combining multi-scale feature extraction and attention mechanisms, GLIA-Net performs excellently in various medical image segmentation tasks, especially when dealing with complex anatomical structures and targets with unclear boundaries, demonstrating significant advantages.
Table 2 presents a performance comparison of various models on internal and external test sets for intracranial aneurysm segmentation. The metrics used include Precision, recall, Dice similarity coefficient (DSC), 95% Hausdorff distance (HD95), area under curve (AUC), and Average Precision (AP). Noting that the evaluation metrics in the table are defined in Reference17.
On the internal test set, GLIA-Net and MGLA-Net significantly outperform U-Net and HeadXNet. However, the proposed method excels in HD95 (4.51), suggesting a more precise segmentation boundary and fewer outliers compared to GLIA-Net (9.07). While MGLIA-Net has a slightly lower recall (70.5%) on the internal set, its improved Precision (52.9%) and AUC (98.9%) indicate a better balance between precision and recall.
In the external test set A, this trend continues. GLIA-Net achieves an accuracy of 71.2%, a recall of 83.9%, and a Dice coefficient of 76.8%, while MGLA-Net scores 72.1% accuracy and 82.5% recall. This consistent performance suggests that these models generalize well to unseen data. The 95% Hausdorff distance further highlights MGLA-Net’s superior performance (3.07), indicating better boundary delineation. GLIA-Net follows with 8.28, while U-Net and HeadXNet show larger distances, suggesting significant inaccuracies.
The same trend is suggested for set B. Overall, MGLIA-Net and GLIA-Net outperform U-Net and HeadXNet across all metrics, particularly in accuracy and boundary accuracy. The superior performance MGLIA-Net and GLIA-Net likely stems from their incorporation of global context, which both of them leverage global localization information to guide local segmentation. For MGLIA-net, it decouple spatial and channel-wise convolutions, and since separable convolution significantly reduces the amount of computation, deeper networks can be constructed without worrying about the risk of overfitting, and deeper networks can usually capture richer details, thereby improving the segment accuracy of small targets such as intracranial aneurysms.
In order to further carefully verify the performance of a proposed method against GLIA-Net, a baseline method, for segmenting intracranial aneurysms (IAs) of different sizes. The datasets are split into three size categories and the proportion is indicated in the Table 3. The segmentation results of three different shapes of intracranial arterial aneurysm cases were tested below, and three sets of experiments were designed: large, medium and small shape intracranial aneurysm prediction and segmentation experiments.
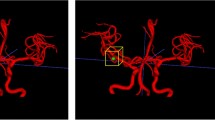
From the qualitative point of view, the visualization results are shown in Figs. 8 and 9, and 10, which respectively display the segmentation results of the cross-sectional planes, sagittal planes, and coronal planes, as well as the enlarged local details graph. The green area represents the true annotation labels, and the red area represents the algorithm’s predicted results. The higher the overlap between the red and green areas, the better the segmentation accuracy. Non overlapping areas were marked with red double arrows in different experimental results.
When comparing the performance of GLIA Net and our method in the segmentation of intracranial aneurysm, experiments were conducted on different shapes of aneurysm. As shown in Fig. 8, for larger intracranial aneurysm, the segmentation results of the two algorithms can coincide well with the actual labels. This indicates that both algorithms could achieve high accuracy when dealing with large-sized aneurysms, because large-sized aneurysms have obvious features and are easy to identify and segment. Figure 9 shows the segmentation results of medium-sized intracranial aneurysm. By observing the overlapping area marked by the red arrow, it can be clearly seen that the results obtained by GLIA Net have a smaller overlapping area and lower accuracy. When further testing small-sized intracranial aneurysm, as shown in Fig. 10, the overlap area between GLIA-Net and the proposed method is relatively small. However, through observation at the arrow mark, it can still be clearly observed that the overlap area of GLIA Net is smaller than that of the proposed method, indicating lower accuracy.
Quantitatively, for small IA Segmentation (Table 4): The proposed method demonstrates a noticeable improvement in Precision compared to GLIA-Net across all three datasets. While Recall is slightly lower for the proposed method the DSC (a measure of overall segmentation overlap) is comparable. Crucially, the 95% HD, which measures the maximum segmentation error, is significantly lower for the proposed method indicating better boundary delineation. This is a key advantage in small IA detection, where accurate boundary definition is critical.
For medium IA Segmentation (Table 5): The performance of both methods is more similar in this size category. The proposed method shows slightly better Precision, while GLIA-Net has slightly better Recall on the Internal and External A sets. DSC and 95% HD are generally comparable, with the proposed method showing lower 95% HD. The AUC and AP values are also close.
For Large IA Segmentation (Table 6): The performance is very similar between the two methods for large IAs. There are no substantial differences in Precision, Recall, DSC, or 95% HD.
The results indicate that the proposed method offers a significant advantage in the segmentation of small intracranial aneurysms (≤ 3 mm), particularly in terms of Precision and 95% Hausdorff Distance. This suggests improved accuracy and more precise boundary detection for small IAs, which are often the most challenging to segment. For medium and large IAs, the performance of the proposed method is generally comparable to that of GLIA-Net. The improved performance on small IAs is a valuable contribution, as accurate detection and segmentation of these aneurysms are crucial for risk assessment and treatment planning.
Conclusions
In this paper, a GLIA-Net algorithm based on MoblieNet (MGLIA-Net) is developed by combining GLIA-Net network. For intracranial aneurysm, a detection target with stereoscopic structure, considering the spatial correlation of images, simultaneously modeling cross channel correlation convolution, better generalization ability is achieved. In order to verify the effectiveness of the algorithm in intracranial aneurysm segmentation, performance tests were carried out on different open source datasets. The results show that the segmentation accuracy of the proposed algorithm reaches 52.9%, 72.1% and 60.1% respectively on three different data sets, which is significantly better than the original GLIA-Net algorithm. This provides a reliable data foundation for more accurate 3D reconstruction in the future, and in the analysis of blood fluid stress, accurate segmentation results can more accurately simulate the blood flow state, which is conducive to further exploring the formation and development mechanism of intracranial aneurysms.
Data availability
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
References
Hoh, B. L. et al. 2023 Guideline for the management of patients with aneurysmal subarachnoid hemorrhage: a guideline from the American heart association/american stroke Association[J]. Stroke 54 (7), e314–e370 (2023).
Neifert, S. N. et al. Aneurysmal subarachnoid hemorrhage: the last decade[J]. Translational Stroke Res. 12, 428–446 (2021).
Brisman, J. L., Song, J. K. & Newell, D. W. Cerebral aneurysms[J]. N. Engl. J. Med. 355 (9), 928–939 (2006).
Abdulazim, A. et al. Diagnosis of delayed cerebral ischemia in patients with aneurysmal subarachnoid hemorrhage and triggers for intervention[J]. Neurocrit. Care. 39 (2), 311–319 (2023).
Szántó, D. et al. Diagnosis and management of Takotsubo syndrome in acute aneurysmal subarachnoid hemorrhage: A comprehensive Review[J]. Rev. Cardiovasc. Med. 24 (6), 177 (2023).
Thilak, S. et al. Diagnosis and management of subarachnoid haemorrhage[J]. Nat. Commun. 15 (1), 1850 (2024).
Bechan, R. S. et al. CT angiography versus 3D rotational angiography in patients with subarachnoid hemorrhage[J]. Neuroradiology 57, 1239–1246 (2015).
Philipp, L. R. et al. Comparison between CTA and digital Subtraction angiography in the diagnosis of ruptured aneurysms[J]. Neurosurgery 80 (5), 769–777 (2017).
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks[C]//Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6–12, 2014, Proceedings, Part I 13. Springer International Publishing, : 818–833. (2014).
Stember, J. N. et al. Convolutional neural networks for the detection and measurement of cerebral aneurysms on magnetic resonance angiography[J]. J. Digit. Imaging. 32, 808–815 (2019).
He, K. et al. Deep residual learning for image recognition[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. : 770–778. (2016).
Shimada, Y. et al. Incidental cerebral aneurysms detected by a computer-assisted detection system based on artificial intelligence: a case series[J]. Medicine 99 (43), e21518 (2020).
Ueda, D. et al. Deep learning for MR angiography: automated detection of cerebral aneurysms[J]. Radiology 290(1), 187–194 (2019).
Park, A. et al. Deep learning–assisted diagnosis of cerebral aneurysms using the HeadXNet model[J]. JAMA Netw. Open. 2 (6), e195600–e195600 (2019).
Bo, Z. H. et al. Toward human intervention-free clinical diagnosis of intracranial aneurysm via deep neural network[J]. Patterns 2(2), 100197 (2021).
Sandler, M. et al. Mobilenetv2: Inverted residuals and linear bottlenecks[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. : 4510–4520. (2018).
Ronneberger, O., Fischer, P. & Brox, T. U. N. Convolutional Networks for Biomedical Image Segmentation[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing, (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Ronen, R. et al. GLASS: global to local attention for Scene-Text Spotting[J]. (2022). https://doi.org/10.48550/arXiv.2208.03364
Funding
This work was supported by JiLin Science and Technology plan project (20220401089YY, 20210401143YY).
Author information
Authors and Affiliations
Contributions
Xin Feng: study concept and design, critical revision of manuscript for intellectual content. Ke Meng : acquisition of data and clinical diagnosis and analysis. LiJie Hou: collect and analysis data, and write manuscripts. Jian Zhang and Lihui Zhao: acquisition and interpretation of data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hou, L., Zhang, J., Zhao, L. et al. CTA image segmentation method for intracranial aneurysms based on MGLIA net. Sci Rep 15, 10593 (2025). https://doi.org/10.1038/s41598-025-95143-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-95143-2