Abstract
Porcine transmissible enteritis virus (TGEV) is a fatal pathogen affecting newborn piglets, presenting a significant challenge to global intensive pig farming biosecurity due to its ongoing mutation. There is still a lack of effective vaccines to combat this virus, Vaccination has long been considered the most effective way to overcome infectious diseases, however, traditional vaccines cannot be brought to market quickly enough to deal with rapid mutations and emerging viruses. Therefore, this study addresses this gap by using immunoinformatics methods and ferritin nanoparticle delivery system to build a platform for rapid research and development of porcine coronavirus vaccine, designing a candidate nanoparticle vaccine that targets the TGEV S protein. To this end, multiple servers and strict screening criteria were used to analyze the S protein, and 3 CTL dominant epitopes, 3 Th dominant epitopes, and 6 B cell dominant epitopes were obtained. The candidate nanoparticle vaccine was constructed by incorporating ferritin sequences through the C-terminus after they were tandemly linked in a certain order using a flexible linker. Further experimental analyses showed that the designed candidate nanoparticle vaccine possessed relatively high antigenicity, immunogenicity, non-allergenicity, non-transmembrane proteins, suitable physicochemical properties, and high solubility upon overexpression. Tertiary structure modeling and disulfide engineering ensured conformational similarity to natural proteins and high stability. Additionally, the model predicted 6 Linear Epitopes and 6 Discontinuous Epitopes for B-cell conformational epitopes. Docking with TLR-3 and TLR-4 molecules shows a large number of interacting hydrogen-bonded amino acid residues and hydrophobically interacting amino acid residues. Immunomimetic assays show high levels of immunoglobulin, T-lymphocyte and IFN-γ secretion and may elicit specific immune responses. Through computerized cloning, the candidate nanoparticle vaccine can be efficiently expressed in the E. coli K12 expression system, aligning with future large-scale industrial production strategies. Overall, the results indicate that the constructed candidate nanoparticle vaccine can be effectively expressed and may be able to induce a strong immune response, which is expected to be an ideal candidate vaccine against TGEV.
Similar content being viewed by others
Introduction
Transmissible gastroenteritis (TGE) is a highly contagious acute digestive system disease caused by transmissible gastroenteritis virus (TGEV) infection1. This infection manifests in viral enteritis, severe watery diarrhea, loss of appetite, vomiting, dehydration and often, high mortality. Pigs of different ages can be infected with TGEV, especially piglets under two weeks of age are the most susceptible to infection, with mortality rates as high as 100%2. This disease has become a worldwide epidemic and is one of the important diseases that cause early death of piglets in various pig-raising countries, causing heavy economic losses to the global pig breeding industry3. Currently, there are no effective drugs for treating TGEV infection, and vaccination is considered to be the most effective strategy for control and prevention, whereby the vaccine stimulates the body to develop an immune response and produce protective antibodies, which in turn produce antiviral functions4. Conventional vaccines (attenuated vaccines, inactivated vaccines or triple vaccines) are mostly used in China, but they have defects such as unreliable immune effects and atavism5. Due to its frequent genetic variation, the circulating strain does not match the vaccine strain, and vaccine development lags behind2, Therefore, an efficient and rapid development platform combined with a broad spectrum vaccine development strategy will hopefully solve this problem.
TGEV is an enveloped, single-stranded positive RNA virus with a full genome length of approximately 28.5 kb organized into a 5′UTR-ORF1a/lb-S-3a-3b-E-M-N-ORF7-3′UTR structure, containing nine open reading frames (ORFs), with ORF2, ORF4, ORF5 and ORF6 encoding four structural proteins, namely, Spike (S), Nucleocapsid (N), Membrane (M), Envelope glycoprotein (E) respectively6. Among these, the S protein is the main structural protein of TGEV, located on the virus surface, functioning as the main antigen inducing the body to produce neutralizing antibodies, and plays an important biological role in virus virulence, histophilicity, receptor-binding sites, and hemagglutination, which is the main target for vaccine development7,8. However, full-length S proteins pose challenges due to potential antigenic interference and toxicity concerns when used in vaccine formulations9. In addressing this challenge and achieving immune homeostasis at the same time, studies have used immunoinformatics tools to examine the T- and B-cell dominant epitopes of the S-protein. Combining computer science and immunology, this approach leverages advanced neural networks to screen S gene sequences using various algorithms thoroughly, elucidating key epitope information for vaccine design10. Yet epitope-based vaccines encounter difficulty in eliciting a robust immune response, necessitating delivery platforms to enhance antigen immunogenicity11. Considering ferritin’s widespread use as a nanoparticle delivery platform, its 24 subunits are arranged symmetrically around a hollow core in an octahedral manner, providing a highly ordered, repetitive structure with abundant surface area for multivalent display of target sequences to enhance immune responses and vaccine effectiveness12, this study explored the tandem fusion of ferritin’s N-terminal end with epitopes to construct candidate nanoparticle vaccine. These candidate nanoparticle vaccine were constructed with a view to providing a practical new approach for the creation of new anti-TGEV vaccines and to provide theoretical and data support for the improvement of antigen design in nanoparticle vaccine research.
Materials and methods
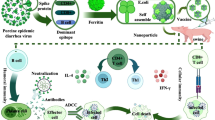
The research employs theoretical guidance to design candidate nanoparticle vaccines, primarily involving the screening of T-cell and B-cell dominant epitopes. Ferritin self-assembled nanoparticles serve as the delivery system for constructing these candidate nanoparticle vaccines. Subsequently, the vaccine properties are analyzed, followed by immune simulation and computerized cloning. As shown in Fig. 1 (Draw with the “Biorender” website), the immune response mechanism caused by the candidate nanoparticle vaccine is briefly described.
T cell epitope prediction
The TGEV-S protein cellular epitope prediction template sequence (GenBankID: ACN71196.1) was acquired in FASTA format from the NCBI database. T cell epitopes are divided into cytotoxic T cell epitopes (CTL) and helper T cell epitopes (Th). CTL epitopes were analyzed using the NetMHCpan4.1 server (https://services.healthtech.dtu.dk/service.php? NetMHCpan-4.1) and the NetMHCpanEL4.1 server (http://tools.iedb.org/mhci/)13,14. Both servers use the same screening method, employing the consensus recommendation method composed of ANN, SMM, and CombLib. These methods use artificial neural networks to predict the binding of known epitopes to MHC molecules, identifying strong binding sequences (5%) within the same allele as potential candidates for sequences of length 14. Th epitopes were analyzed using the NetMHCIIpan-4.0 server (https://services.healthtech.dtu.dk/service.php? NetMHCIIpan-4.0)15, which also uses an artificial neural network to predict potential binding peptide segments of any known MHC-II molecule sequence and outputs the likelihood of the peptide segment naturally existing with MHC-II alleles, identifying strong binding epitopes (10%) of length 15. MHC-II molecules activate macrophages to recognize and clear intracellular pathogens by releasing IFN-γ. The secretion of cytokine IL-4 promotes the proliferation and differentiation of antigen-presenting cells. Therefore, Epitope evaluation included IFN epitope analysis (https://webs.iiitd.edu.in/raghava/ifnepitope/index.php) and IL4pred (http://crdd.osdd.net/raghava/il4pred/) for IFN⁃γ and IL inducibility16,17.
B cell epitope prediction
Linear B-cell epitopes were analyzed by the ABCpred server (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html)18, a trained recurrent neural network server that grades predicted B-cell epitopes based on their quality. The analysis was conducted with a sequence length set to 16, and a prediction threshold of 0.8 was applied to ensure robustness. Additionally, to ensure high confidence in the results, the entire amino acid sequence was analyzed using the PEPTIDES server (http://imed.med.ucm.es/Tools/antigenic.html)19. The PEPTIDES server employs the Kolaskar and Tongaonkar methods to predict potential antibody responses by simulating 3D epitopes of immunogenicity recognition, reporting fragments only when the minimum number of residues is 8.
Screening and conservativeness analysis of candidate epitopes
After applying the filters above, the outcomes from various servers were compared, focusing on the overlapping regions of epitopes. The immunogenicity of these overlapping epitopes was assessed using the IEDBMHC-I server (http://tools.iedb.org/immunogenicity/)20, while toxicity was evaluated via the ToxinPred server (https://webs.iiitd.edu.in/raghava/toxinpred/ design.php) 21. Epitopes with an immunogenicity score > 0.25 and deemed non-toxic were designated as candidate epitopes. Candidate epitopes were evaluated for conservation with 45 TGEV-S protein amino acid sequences downloaded from the NCBI database via the IEDB server (http://tools.iedb.org/conservancy/)22. At the same time, the conservation of amino acid residues in dominant epitopes is plotted through Weblogo.
Vaccine design and evaluation
CTL, Th, and B cell dominant epitopes were tandem linked using AAY, GPGPG, and KK flexlinker, respectively. AAY provides flexibility to protect proteins from degradation, GPGPG facilitates immunoprocessing and presentation, while KK, a dual lysyl linker, retains epitope-independent immunoreactivity. Ferritin is a self-assembled nanoparticle whose highly ordered and repetitive symmetric structure serves as a nanoplatform for antigen delivery and immunostimulation, contributing to the induction of efficient immune responses and enhancing vaccine efficacy. The N-terminal of ferritin (GenBank: WP_000949190) was linked to the dominant epitope construct via EAAAK to generate a candidate nanoparticle vaccine. The antigenicity of candidate nanoparticle vaccines was assessed using the ANTIGENpro server (http://scratch.proteomics.ics.uci.edu/)23, while solubility was evaluated using the SOLpro overexpression server (http://scratch.proteomics.ics.uci.edu/index.html)24. AllerTop Sensitization Server (http://www.ddg-pharmfac.net/AllerTOP/)25, ProtScale Hydrophobicity Server (https://web.expasy.org/cgi-bin/protscale/protscale.pl#opennewwindow)26, TMHMM-2.0 Transmembrane Structural Domain Server (https://services.healthtech.dtu.dk/services/TMHMM-2.0/) for comprehensive analysis27, and the ExPASy Physicochemical Properties Server (https://web.expasy.org/protparam/) for evaluating various computational parameters including relative molecular mass, theoretical pI, atomic composition, estimated half-life, extinction coefficient, instability index, fat index and GRAVY28.
Vaccine secondary structure prediction and tertiary structure modeling and refinement, disulfide engineering and validation
Vaccine secondary structures were analyzed using the SOPMA server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) and PSIPRED 4.0 server (http://bioinf.cs.ucl.ac.uk /psipred) to analyze transmembrane helices, folding and domain identification29,30. Tertiary structure modeling was conducted using the homology modeling trRosetta server (https://yanglab.nankai.edu.cn/trRosetta/) to generate primary 3D models31. Subsequently, the primary 3D model pdb files were subjected to molecular dynamics simulations performing repetitive structural perturbations32 through the GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?), aimed at refining errors in the primary initial models. The refined model was further stabilized by disulfide engineering through the DbD2 server (http://cptweb.cpt.wayne.edu/DbD2/index.php) to enhance structural stability33. The models were evaluated before and after refinement using the CHECK Serve (https://saves.mbi.ucla.edu/)34, with the final models subjected to validation through Z-score generation via the ProSA-web server (https://prosa.services.came.sbg.ac.at/prosa.php)35.
Analysis of conformational B-cell epitopes and molecular docking
Conformational B-cell epitopes, comprising both discontinuous and continuous epitopes formed through protein folding, were evaluated in the final refined candidate nanoparticle vaccine tertiary structure using the ElliPro server (http://tools.iedb.org/ellipro/)36. A threshold score > 0.5 and a maximum distance of 6 Angstrom were employed to assess the presence of these epitopes. Also, in order to properly stimulate an immune response, interactions between antigenic and immune receptor molecules are essential. TLR3, known for regulating immune responses, promoting immune cell maturation and differentiation, and participating in pro-inflammatory factor responses, plays a vital role in intrinsic antiviral immune responses. Conversely, TLR4 acts as a key pattern recognition receptor, inducing cytokines and bridging innate and acquired immunity, contributing to the host cell’s resistance against viral infections. Protein–protein docking studies were conducted using the pyDockWEB server (https://life.bsc.es/pid/pydockweb) to analyze the interaction patterns between the candidate nanoparticle vaccine and TLR-3 (PDB ID: 2A0Z) and TLR-4 (PDB ID: 4G8A)37. Docking complexes were visualized using the PyMOL tool, while complex 2D interworking interfaces were visualized by the LigPLot + tool.
Molecular dynamics simulation
Molecular dynamics simulations were performed using the Gromacs 2022.3 software to examine the stability of the candidate vaccine docking complex38,39. The GAFF force field and hydrogenation operation were added to each complex and RESP potential was calculated. The Tip3p water model was used for step energy minimization and balance protocol. The simulation environment of 300 K and 1 Bar was selected and the trajectory analysis was carried out in 100 ns time to calculate the physical quantities of RMSD, RMSF, Rg, SASA and hydrogen bond of the complex. In addition, the Gibbs free energy topography was evaluated by using the built-in “gmx_sham” function and "xpm2txt.py" foot.
Immunosimulation and computerized cloning
The immune simulation of the candidate nanoparticle vaccine was performed using the C-ImmSim server (https://150.146.2.1/C-IMMSIM/index.php)40, an agent-based computational immune response simulator that utilizes position-specific scoring matrices (PSSM) and machine learning methods, respectively, to predict epitope immune interactions. To mimic commercial vaccine dosing intervals, the immunization simulation retained default parameters, with the number of simulation steps set to 1050 and time steps adjusted to 1, 84, and 170 (8 h per step) for three immunizations. In order to express foreign genes in host organisms, codon optimization is necessary depending on the particular host organism; therefore, candidate nanoparticle vaccine sequences were submitted to the JCat server (http:/jcat.de/)41 for codon optimization using E. coli K12 as the host. ExpOptimizer server (https://www.novopro.cn/tools/codon-optimization.html) (NOVOPRO, China) validated Codon Adaptation Index (CAI) values against GC content. Finally, the adapted nucleotide sequences were cloned into the pET28-a( +) expression vector using SnapGene software.
Result
Results of the TGEV S protein dominant epitope screening
From the 1448 amino acids of S protein obtained from NCBI (The complete amino acid sequence is shown in supplemental table S1), CTL dominant epitopes were identified using the NetMHCpan-4.1 and NetMHCpanEL4.1 dual servers, revealing three overlapping dominant epitope regions. The dominant epitopes were analyzed via the NetMHCIIpan-4.0 server and further evaluated by IFNepitope and IL4pred server, identifying three overlapping dominant epitope regions. Screening with the ABCpred and PEPTIDES servers revealed six overlapping linear B-cell epitope regions. All identified dominant epitopes were confirmed as non-toxic peptides by the ToxinPred server and highly immunogenic (immunogenicity score > 0.25) by the IEDBMHC-I server. Conservation analysis conducted by the IEDB server indicated high conservation of linear B-cell dominant epitopes with Th-2 and Th-3 across all 45 sequences (The S protein GenBankID of 45 strains was shown in supplemental table S2), while CTL dominant epitopes showed less conservation with Th-1 (Detailed scores for conservative analysis are shown in supplementary table S3). Nonetheless, considering their significance in vaccine molecular design, they were included in the study. The results are shown in Table 1. The analysis of amino acid residues using Weblogo showed that most of the dominant epitopes were conserved, with only a few amino acid residues experiencing mutations, as shown in Fig. 2. In the analysis of epitope hiding, it was found that all the selected epitopes except B-6 epitopes were located externally (The complete extracellular region of S protein is shown in supplementary figure S8).
TGE S protein vaccine construction
Based on the above analytical screening, the candidate nanoparticle vaccine was designed by selecting the above T-cell and B-cell potential epitopes for vaccine construction. Figure 3 illustrates the selection of 12 dominant epitopes. Th epitopes were connected using GPGPG flexlinker, CTL epitopes using AAY flexlinker, and B-cell epitopes using KK flexlinker. Additionally, the ferritin N-terminus was tandemly linked with the C-terminus of the dominant epitope B-6 using EAAAK flexlinker to form the complete candidate nanoparticle vaccine (The amino acid sequence of the candidate vaccine is shown in supplemental Table S4).
Assessment of candidate nanoparticle vaccine
The antigenicity evaluation of the candidate nanoparticle vaccine yielded a score of 0.752474 using the ANTIGENpro server. Solubility analysis during overexpression indicated a score of 0.647714 via the SOLpro server. Additionally, the AllerTop server confirmed the non-sensitizing nature of the candidate nanoparticle vaccine, indicating its antigenicity, solubility, and non-sensitizing properties. ProtScale server analysis of hydrophobicity revealed a large number of negative regions for candidate nanoparticle vaccines, and the results are shown in Fig. 4A. The TMHMM-2.0 server analyzed the transmembrane structural domains in the candidate nanoparticle vaccine were all extracellular regions, and the results are shown in Fig. 4B, indicating that the candidate nanoparticle vaccine has hydrophilic and extracellular secretion functions. Further characterization of physical and chemical properties via the ExPASy server revealed essential parameters: the candidate nanoparticle vaccine comprised 368 amino acids, with a molecular weight of 41814.09 and a theoretical pI of 6.69. The instability index (II) computed as 28.75 classifies the protein as stable—aliphatic index as 91.47 and the Grand average of hydropathicity (GRAVY) as -0.153. The secondary structure analysis, performed using the SOPMA and PSIPRED servers, depicted a composition of Helix, sheet, Turn, and Coil, with PSIPRED indicating percentages of Alpha helix (43.75%), Extended strand (22.01%), Beta turn (8.42%), and Random coil (25.82%), as shown in Fig. 4C,D, respectively.
Results of candidate nanoparticle vaccine evaluation. (A) candidate nanoparticle vaccine hydrophobicity analysis; (B) candidate nanoparticle vaccine transmembrane domain analysis; (C) candidate vaccine secondary structure analyzed by SOPMA server; (D) candidate vaccine secondary structure analyzed by PSIPRED server.
Modeling, refinement and evaluation of tertiary structures of candidate nanoparticle vaccine
The tertiary structure of the candidate nanoparticle vaccine was generated using the trRosetta server via homology modeling, resulting in a model with a TM-score of 0.632. Visualization of the structure was performed using Pymol software, as depicted in Fig. 5F. Additionally, the trRosetta server generated a 2D view of the tertiary structure, and the results are shown in Fig. 5A–E. To ensure that the protein resembled the natural structure, the original tertiary structure model of the generated candidate vaccine was later refined using the Galaxy-Refine server (The detailed scores of the tertiary structure refinement model are shown in supplementary table S5). Structural comparisons were made using Pymol software, with results illustrated in Fig. 6B. The level of refinement pre- and post model refinement was assessed by generating Ramachandran plots of the protein’s tertiary structure using the PROCHECK server. In the crude model, 88.4% of residues fell within the most favoured regions, 10.0% in additional allowed regions, 0.6% in generously allowed regions, and 0.9% in disallowed regions, as depicted in Fig. 6A. Following refinement, the percentages shifted to 93.6%, 4.9%, 0.9%, and 0.6% for residues in the respective regions, as illustrated in Fig. 6C (The detailed comparison results before and after the tertiary structure refinement of candidate vaccines are shown in supplementary Figure S1-S5). The results demonstrate that the model quality was improved after refinement. However, further stabilization of the protein tertiary structure through disulfide engineering was deemed necessary for the refined model. This engineering was executed via the DbD2 server. Notably, the refined model displayed a B-factor of 0 in the analysis results, indicating that no sequence mutations were required to introduce disulfide bonds, as shown in Fig. 6D. The ProSA web server performed the final refined model scoring and validation, and the results are shown in Fig. 6E,F. The model exhibited a Z-Score of -4.83, followed by predominantly negative local energy scores. These findings affirm that the refined model conforms to natural protein conformation, rendering it an optimal candidate for subsequent analytical steps.
Modeling of the tertiary structure of the candidate nanoparticle vaccine. (A)–(E): Are 2D structural parameters (including Omega, Phi, Distance, Theta, and Contact); (F) The tertiary structure of the candidate nanoparticle vaccine as visualized by the Pymol software (Th-dominant epitopes are in blue, CTL-dominant epitopes are in purple, B-cell-dominant epitopes are in yellow, and ferritin is in red).
Refinement of the tertiary structure of the candidate nanoparticle vaccine. (A) Ramachandran graphic validation of the tertiary structure; (B) cartoon comparison of the before and after model structure (green for the coarse model, red for the refined model); (C) Ramachandran graphic validation of the refined tertiary structure; (D) disulfide engineering of the refined model (the yellow part is the disulfide bond); (E) ProSA SEB server Evaluating the refined model; (F) ProSA SEB server evaluating the local model quality.
Analysis of structural B cell epitopes
Ellipro server identified a total of 6 Linear epitopes and 6 discontinuous epitopes within the 368 amino acids of the candidate nanoparticle vaccine. As presented in Table 2, all predicted scores surpassed the server’s threshold of 0.5.
Docking of candidate nanoparticle vaccine molecules
The ability of candidate nanoparticle vaccines to interact with TLRs is an important indication of defense against pathogens. To evaluate this, protein–protein docking was conducted using the pyDockWEB server. The server comprehensively ranked the complexes, with the top-ranked docked complexes selected for subsequent analysis. From the docking analysis, it is obvious that the candidate nanoparticle vaccine can be stably docked with TLR-3 and TLR-4. The composite scores (kcal/mol) for docking with TLR-3 were as follows: Electrostatics: -10.549, Desolvation: -47.666, Vdw: 86.453, with a Total score of -49.569. Pymol software was utilized to visualize the local interaction sites, as depicted in Fig. 7A–C, while LigPlot + software generated the 2D interaction interface, illustrated in Fig. 7D, There were 6 amino acid residues with hydrogen bond and 23 amino acid residues with hydrophobic interaction. The composite scores (kcal/mol) for docking with TLR-4 were as follows: Electrostatics: -9.895, Desolvation: -47.614, Vdw: 63.102, resulting in a Total score of -51.199. Pymol software was used to visualize the local interaction sites, depicted in Fig. 7E–G, while LigPlot + software generated the 2D interaction interface, illustrated in Fig. 7H, There were 6 amino acid residues with hydrogen bond and 23 amino acid residues with hydrophobic interaction. Overall, the docking analysis revealed total energy consumption scores around − 50 kcal/mol, with the two-dimensional interaction interface displaying numerous interacting amino acid residues, indicating a robust interaction between the candidate vaccine and TLR-3 and TLR-4.
Results of molecular docking of candidate nanoparticle vaccine with TLR-3 and TLR-4. (A) and (E): Docking of candidate nanoparticle vaccine with TLR-3 and TLR-4 (candidate vaccine structure is in red, TLR-3 and TLR-4 are in green); (B) and F: Magnification of local interaction position; (C) and (G) Specific interaction amino acid residues; (D) and (H): Two-dimensional interaction interface.
Molecular dynamics simulation analysis
In MD simulation, Fig. 8A–G shows the results of docking complex analysis between TLR-3 and candidate vaccine, and Fig. 8H–N shows the results of docking complex analysis between TLR-4 and candidate vaccine. It can be seen from the figure that RMSD curves remain between 4–5 nm and 12–14 nm respectively after 45 ns, indicating that the complex tends to be stable during the simulation process and the structure is relatively stable. The RMSF curve Chain A showed different behaviors up to 1.5 and 4.0 nm, respectively. Most of the regions in TLR-3 showed relative stability, while some regions in TLR-4 showed higher fluctuations, which proved that the dynamic behavior of amino acid residues was more stable in the TLR-3 complex. Rg curve analysis further demonstrated the spatial conformation of the complex, and both complexes fluctuated within 100 ns. In the analysis of hydrogen bond curves, about 10 and 20 hydrogen bonds were found in 20 to 60 ns, respectively, showing strong interactions. SASA analysis showed a decline from 560 to 500 nm2 and 810 nm2 to 750 nm2 within the first 40 ns, respectively, and then remained stable, reflecting the tight binding and stability of the complex. In Gibbs free energy analysis, the 3D free energy landscape map is drawn, and the RMSD is in the range of 0.0–2.5. The Gyrate fell to 0.0–4.8 and 0.0–5.8 respectively, which further confirmed the relatively good stability of the two docking complexes.
Immune simulation of candidate nanoparticle vaccine
The immune simulation reaction was performed on the C-ImmSimm server, and the results are shown in Fig. 9. The candidate nanoparticle vaccine was injected three times and significantly induced primary immune responses after three injections. And three peak curves were formed, which proved that immune memory was successfully created and memory T and B lymphocytes produced a rapid response. Notably, secondary immune responses were stimulated, with gradual increments observed in primary immune responses after each subsequent injection. A marked elevation in secondary and tertiary antibody levels (IgG1 + IgG2, IgM and IgG + IgM) was observed. Furthermore, there was a continual increase in the concentration of active B cells, plasma B cells, helper T cells, and cytotoxic T cells, peaking during antigen injection, indicating effective immune responses, establishment of immune memory, and efficient antigen clearance. Increases in dendritic cells and macrophages pointed to excellent antigen presentation by antigen-presenting cells (APCs), while the candidate nanoparticle vaccine resulted in significant increases in cytokine (IFN-γ, IL-4, and IL-10) levels. Therefore, the simulated immune response demonstrated that the candidate nanoparticle vaccine elicited substantial production of immunoglobulins, APCs, cytokines, and activated B and T cells, showcasing its ability to induce potent immunogenic responses post-vaccination within the host.
Results of immune response induced by three doses of the vaccine. (A) Overall change trend of B lymphocytes; (B) Plasma B lymphocytes count; (C) B lymphocytes population per entity-state; (D) and (E) Helper t lymphocyte population; (F) Regulatory T cell population; (G) Cytotoxic T lymphocyte population; (H) Cytotoxic T-cell population per state; (I) Natural killer cell population; (J) Dendritic cell population per state; (K) Macrophages population per state; (L) EP cell population; (M) The change trend of antigen and immunoglobulin; (N) Trends of cytokines and interleukin.
Codon optimization and in-silico cloning of vaccine
The efficient expression of candidate nanoparticle vaccines is a key step in silic cloning. Therefore, the amino acid sequences of the candidate nanoparticle vaccine were codon-optimized for prokaryotic organisms to improve their translational efficiency based on E. coli K12 in the JCat server. The full length of the nucleotide sequence of the candidate nanoparticle vaccine was 1104 bp after reverse translation (The optimization results of Escherichia coli favoritism are shown in supplementary Figure S6, The complete nucleotide sequence is shown in supplemental table S6). The CAI-Value of the improved sequence reached 1.0, while the GC-Content of E coli was at 47.92%. Upon validation using the ExpOptimizer server, the CAI was determined to be 0.74, with a corresponding GC content of 47.92%. These results, presented in Fig. 10A,B, indicated that all CAI values exceeded the threshold of 0.5, ensuring abundance in the DNA sequences. Additionally, the GC content fell within the optimal scoring range of 30 to 70%. To facilitate expression and purification, XhoI and BamHI restriction endonuclease sites were selected using SnapGene software, and the adapted nucleotide sequences were inserted into the pET28-a(+) expression vector with a 6× his tag sequence. The prokaryotic expression plasmids were constructed, and the results are shown in Fig. 10C. To verify successful insertion, the constructed plasmids underwent double enzyme digestion and PCR simulation, followed by analysis using 1% agarose gel electrophoresis. The results, shown in Fig. 10D,E, confirmed the successful insertion of the candidate vaccine, with bands observed at the correct positions (See supplementary Figure S7 for the complete agarose gel electrophoresis simulation).
Construction of the prokaryotic expression system of the candidate nanoparticle vaccine. (A) ExpOptimizer server to verify the CAI value; (B) ExpOptimizer server to verify the GC content; (C) Candidate nanoparticle vaccine (red) inserted into pET-28a(+); (D) Double enzymatic digestion of pET-28a-Vaccine plasmid (Cropped from supplemental Figure S7); (E) PCR simulation of the candidate nanoparticle vaccine Analysis (Cropped from supplemental Figure S7).
Discussion
In recent years, coronaviruses have caused serious public health problems with an alarming frequency of recombination and mutation, posing a great threat to human and animal health42. Porcine enteric coronaviruses have led to more frequent infections in swine herds and have caused significant losses to the global farming industry due to their potential for cross-species transmission1. Transmissible gastroenteritis virus causes acute, high-contact enteric infections in neonatal piglets, which are often clinically mixed with other enteroviruses or secondary infections, posing a serious challenge to disease prevention and control in swine farms3. Vaccination is known as a highly effective public health measure for disease prevention, aiding in immune response stimulation and pathogen containment, yet the scarcity of approved and efficacious viral vaccines remains a problem4. At present, many achievements in basic virology, vaccine technology, immunoinformatics and synthetic biology have opened up new opportunities for vaccine development against TGEV43. Strategies for the development of new vaccines as appropriate solutions for the prevention and treatment of viral infections are a primary need and will help to find solutions to the current biosecurity problems on swine farms.
However, the preparation and development of new vaccines is a complex and systematic process constrained by uncertainties related to safety, efficacy, and production feasibility, which limits the advancement of traditional vaccinology and the widespread use of existing vaccines44. One promising method to overcome these challenges is the development of subunit vaccines using immunoinformatics, which relies on advanced computational prediction of protein information45. However, discrepancies between predicted and actual results, along with the intricate and dynamic nature of the animal immune system, can lead to failures in later-stage experiments46. An effective strategy to mitigate these issues is to perform multiple server comparison screenings for the same sequence and use different alleles of animals for a comprehensive evaluation47. Since the epitope of TGEV S protein has not been fully resolved, predictive screening is a necessary method to rapidly enrich its epitope landscape and discard redundant sequence interference. In this study, immunoinformatics-related servers and software were employed for repeated comparison, and screening of TGEV S proteins, and the dominant epitopes identified were combined with a ferritin self-assembled nanoparticle delivery platform to construct candidate vaccines. This tandem polyvalent presentation of dominant epitopes enhances the activation of antigen-presenting cells (APCs) and the maturation of dendritic cells (DCs), thereby amplifying B cell signaling and effectively triggering both humoral and cellular immune responses, It will be beneficial for candidate nanoparticle vaccines to produce protection against TGEV5. Ultimately boosting the host’s protective immunity, the relatively conserved dominant epitope reduces the antigen sequence, alleviating excessive antigen load and allergic reactions of the host, without reversing viral pathogenesis and effectively combating viral genetic variation and antigen drift. The dominant Th-1 and CTL-1 epitopes in the study overlap with the reported neutralizing antibody sites at 538–591 aa and 378–395 aa at site D, providing robust support for the vaccine’s practical efficacy in virus neutralization48. Additionally, to ensure enhanced immune response and higher affinity, the candidate nanoparticle vaccine underwent systematic evaluation (antigenicity, allergenicity, immunogenicity, hydrophobicity, solubility, and toxicity). Most of the secondary structure converted to antigen mosaicism, forming a solid structural basis, with B-cell epitope predictions exceeding server thresholds, indicating a potential for inducing antibody formation49. The docking of TLR-3 and TLR-4 molecules revealed numerous interacting amino acid residues and low binding free energy, MD simulation shows that the docking complex and its conformation are relatively stable at the microscopic level, elucidating the molecular mechanism of antiviral activity and suggesting that the vaccine may induce transcriptional up-regulation of type I interferon, pro-inflammatory cytokines, and chemokines, thereby promoting the host’s antiviral response50. In the immune simulations, Although C-ImmSimm server cannot simulate the ferritin self-assembly process, the secondary and tertiary immune responses following vaccination with candidate nanovaccines exhibited higher levels compared to primary immune responses. This resulted in elevated immunoglobulin production, IFN-γ secretion, and sustained cellular and CTL responses. These findings align with previous observations, such as the results seen in the simulation of COVID-19 S protein47, the research results of tuberculosis vaccine MP3RT in animal experiments51, and the self-assembly results of ferritin and IAV main proteins can trigger a powerful immune response52. Moreover, to address the challenges associated with protein expression purification for large-scale industrial vaccine production, the sequence was incorporated into the pET28-a(+) expression vector with His tags added at both ends. However, despite the systematic design and evaluation of the constructed nanoparticle vaccine candidate, the study’s limitations necessitate future focus on in vitro and in vivo testing to ascertain its practical application.
Conclusions
Multi-epitope vaccines and nanoparticle vaccines have received much attention and progressed to clinical trials. In this study, we successfully combined the two using immunoinformatics leveraging a comprehensive analysis of the S protein’s full-length gene sequence to identify immunologically superior B-cell and T-cell epitopes tandemly linked with ferritin. The resulting candidate nanoparticle vaccine exhibits high antigenicity, along with non-allergenic and non-toxic properties. Through stable binding to immune receptors, TLR-3 and TLR-4, it triggers strong cellular and humoral immunity in the body as shown in the immune simulation and efficient expression in computerized clones. We believe that the candidate nanoparticle vaccine could be a potent vaccine against the TGEV pathogen and, at the same time, provide a new approach and an efficient research platform for accelerating the creation of new vaccines against porcine enteric coronaviruses.
Data availability
The datasets generated and/or analyzed during the current study are included in the text.
Abbreviations
- TGEV:
-
Transmissible gastroenteritis virus
- ORFs:
-
Open reading frames
- Spike protein:
-
S protein
- CTL:
-
Cytotoxic T lymphocyte
- HTL:
-
Helper T lymphocyte
- MHC:
-
Major histocompatibility complex
- IL:
-
Interleukin
- IFN-γ:
-
Interferon gamma
- TLR:
-
Toll like receptors
- pI:
-
Theoretical isoelectric point
- Ig:
-
Immunoglobulin
- CAI:
-
Codon adaptation index
- GRAVY:
-
Grand average of hydropathicity index
- APC:
-
Antigen presenting cell
- DC:
-
Dendritic cells
- NK:
-
Natural killer cells
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- Rg:
-
Radius of gyration
- SASA:
-
Solution accessible surface area
References
Yan, Q. et al. Swine enteric coronavirus: Diverse pathogen–host interactions. Int. J. Mol. Sci. 23(7), 3953. https://doi.org/10.3390/ijms23073953 (2022).
Chen, Y. et al. Transmissible gastroenteritis virus: An update review and perspective. Viruses 15(2), 359. https://doi.org/10.3390/v15020359 (2023).
Pascual-Iglesias, A. et al. Recombinant chimeric transmissible gastroenteritis virus (TGEV)—Porcine epidemic diarrhea virus (PEDV) virus provides protection against virulent PEDV. Viruses 11(8), 682. https://doi.org/10.3390/v11080682 (2019).
Turlewicz-Podbielska, H. & Pomorska-Mól, M. Porcine coronaviruses: Overview of the state of the art. Virol. Sin. 36(5), 833–851. https://doi.org/10.1007/s12250-021-00364-0 (2021).
Du, P. et al. Virus-like particle vaccines with epitopes from porcine epidemic virus and transmissible gastroenteritis virus incorporated into self-assembling ADDomer platform provide clinical immune responses in piglets. Front. Immunol. 14, 1251001. https://doi.org/10.3389/fimmu.2023.1251001 (2023).
Jin, Y. B. et al. Immune responses induced by recombinant Lactobacillus plantarum expressing the spike protein derived from transmissible gastroenteritis virus in piglets. Appl. Microbiol. Biotechnol. 102(19), 8403–8417. https://doi.org/10.1007/s00253-018-9205-0 (2018).
Suo, S. et al. Phage display for identifying peptides that bind the spike protein of transmissible gastroenteritis virus and possess diagnostic potential. Virus Genes 51(1), 51–56. https://doi.org/10.1007/s11262-015-1208-7 (2015).
Trincone, A. & Schwegmann-Weßels, C. Looking for a needle in a haystack: Cellular proteins that may interact with the tyrosine-based sorting signal of the TGEV S protein. Virus Res. 202, 3–11. https://doi.org/10.1016/j.virusres.2014.11.029 (2015).
Golob, J. L., Lugogo, N., Lauring, A. S. & Lok, A. S. SARS-CoV-2 vaccines: a triumph of science and collaboration. JCI Insight. 6(9), e149187. https://doi.org/10.1172/jci.insight.149187 (2021).
Ramana, J., & Mehla, K. Immunoinformatics and epitope prediction. In Methods in Molecular Biology (Clifton, NJ), Vol. 2131, 155–171. https://doi.org/10.1007/978-1-0716-0389-5_6 (2020).
Jaan, S. et al. Multi-epitope chimeric vaccine designing and novel drug targets prioritization against multi-drug resistant Staphylococcus pseudintermedius. Front. Microbiol. 13, 971263 (2022).
Boyoglu-Barnum, S. et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592(7855), 623–628. https://doi.org/10.1038/s41586-021-03365-x (2021).
Trolle, T. et al. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 31(13), 2174–2181. https://doi.org/10.1093/bioinformatics/btv123 (2015).
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48(W1), W449–W454. https://doi.org/10.1093/nar/gkaa379 (2020).
Andreatta, M. et al. An automated benchmarking platform for MHC class II binding prediction methods. Bioinformatics 34(9), 1522–1528. https://doi.org/10.1093/bioinformatics/btx820 (2018).
Dhanda, S. K., Vir, P. & Raghava, G. P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 8, 30. https://doi.org/10.1186/1745-6150-8-30 (2013).
Dhanda, S. K., Gupta, S., Vir, P. & Raghava, G. P. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013, 263952. https://doi.org/10.1155/2013/263952 (2013).
Saha, S. & Raghava, G. P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65(1), 40–48. https://doi.org/10.1002/prot.21078 (2006).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41(Web Server Issue), W349–W357. https://doi.org/10.1093/nar/gkt381 (2013).
Calis, J. J. et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 9(10), e1003266. https://doi.org/10.1371/journal.pcbi.1003266 (2013).
Gupta, S. et al. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8(9), e73957. https://doi.org/10.1371/journal.pone.0073957 (2013).
Dhanda, S. K. et al. IEDB-AR: Immune epitope database-analysis resource in 2019. Nucleic Acids Res. 47(W1), W502–W506. https://doi.org/10.1093/nar/gkz452 (2019).
Cheng, J., Randall, A. Z., Sweredoski, M. J. & Baldi, P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 33(Web Server Issue), W72–W76. https://doi.org/10.1093/nar/gki396 (2005).
Magnan, C. N., Randall, A. & Baldi, P. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics 25(17), 2200–2207. https://doi.org/10.1093/bioinformatics/btp386 (2009).
Dimitrov, I., Bangov, I., Flower, D. R. & Doytchinova, I. AllerTOP vol 2–a server for in silico prediction of allergens. J. Mol. Model. 20(6), 2278. https://doi.org/10.1007/s00894-014-2278-5 (2014).
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D. & Hochstrasser, D. F. Protein identification and analysis tools in the ExPASy server. In Methods in Molecular Biology (Clifton, N.J.), Vol. 112, 531–552. https://doi.org/10.1385/1-59259-584-7:531. (1999).
Ikeda, M., Arai, M., Lao, D. M. & Shimizu, T. Transmembrane topology prediction methods: A re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 2(1), 19–33 (2002).
Duvaud, S. et al. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 49(W1), W216–W227. https://doi.org/10.1093/nar/gkab225 (2021).
Geourjon, C. & Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. CABIOS 11(6), 681–684. https://doi.org/10.1093/bioinformatics/11.6.681 (1995).
Karypis, G. YASSPP: Better kernels and coding schemes lead to improvements in protein secondary structure prediction. Proteins 64(3), 575–586. https://doi.org/10.1002/prot.21036 (2006).
Yang, J. et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 117(3), 1496–1503. https://doi.org/10.1073/pnas.1914677117 (2020).
Heo, L., Park, H. & Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 41(Web Server Issue), W384–W388. https://doi.org/10.1093/nar/gkt458 (2013).
Craig, D. B. & Dombkowski, A. A. Disulfide by Design 2.0: Aa web-based tool for disulfide engineering in proteins. BMC Bioinform. 14, 346. https://doi.org/10.1186/1471-2105-14-346 (2013).
Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8(4), 477–486. https://doi.org/10.1007/BF00228148 (1996).
Lovell, S. C. et al. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 50(3), 437–450. https://doi.org/10.1002/prot.10286 (2003).
Thornton, J. M., Edwards, M. S., Taylor, W. R. & Barlow, D. J. Location of “continuous” antigenic determinants in the protruding regions of proteins. EMBO J. 5(2), 409–413. https://doi.org/10.1002/j.1460-2075.1986.tb04226.x (1986).
Jiménez-García, B., Pons, C. & Fernández-Recio, J. pyDockWEB: A web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics 29(13), 1698–1699. https://doi.org/10.1093/bioinformatics/btt262 (2013).
Sarvmeili, J., Baghban Kohnehrouz, B., Gholizadeh, A., Shanehbandi, D. & Ofoghi, H. Immunoinformatics design of a structural proteins driven multi-epitope candidate vaccine against different SARS-CoV-2 variants based on fynomer [published correction appears in Sci Rep. 14(1), 30524. https://doi.org/10.1038/s41598-024-82829-2.] (2024). Sci. Rep. 14(1), 10297. Published 2024 May 4. https://doi.org/10.1038/s41598-024-61025-2 (2024).
Khatoon, N., Pandey, R. K. & Prajapati, V. K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 7(1), 8285. https://doi.org/10.1038/s41598-017-08842-w (2017).
Rapin, N., Lund, O., Bernaschi, M. & Castiglione, F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 5(4), e9862. https://doi.org/10.1371/journal.pone.0009862 (2010).
Grote, A. et al. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33(Web Server Issue), W526–W531. https://doi.org/10.1093/nar/gki376 (2005).
Liu, Q. & Wang, H. Y. Porcine enteric coronaviruses: An updated overview of the pathogenesis, prevalence, and diagnosis. Vet. Res. Commun. 45(2–3), 75–86. https://doi.org/10.1007/s11259-021-09808-0 (2021).
Chen, Y. et al. Antiviral drugs screening for swine acute diarrhea syndrome coronavirus. Int. J. Mol. Sci. 23(19), 11250. https://doi.org/10.3390/ijms231911250 (2022).
He, J. et al. Vaccine design based on 16 epitopes of SARS-CoV-2 spike protein. J. Med. Virol. 93(4), 2115–2131. https://doi.org/10.1002/jmv.26596 (2021).
Omoniyi, A. A. et al. In silico design and analyses of a multi-epitope vaccine against Crimean-Congo hemorrhagic fever virus through reverse vaccinology and immunoinformatics approaches. Sci. Rep. 12(1), 8736. https://doi.org/10.1038/s41598-022-12651-1 (2022).
Mahmud, S. et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 11(1), 15431. https://doi.org/10.1038/s41598-021-92176-1 (2021).
Sahu, L. K. & Singh, K. Cross-variant proof predictive vaccine design based on SARS-CoV-2 spike protein using immunoinformatics approach. Beni-Suef Univ. J. Basic Appl. Sci. 12(1), 5. https://doi.org/10.1186/s43088-023-00341-4 (2023).
Yuan, X., Lin, H. & Fan, H. Efficacy and immunogenicity of recombinant swinepox virus expressing the A epitope of the TGEV S protein. Vaccine 33(32), 3900–3906. https://doi.org/10.1016/j.vaccine.2015.06.057 (2015).
Meng, F. & Kurgan, L. Computational prediction of protein secondary structure from sequence. In Current Protocols in Protein Science, Vol. 86, 2.3.1–2.3.10. https://doi.org/10.1002/cpps.19 (2016).
Aziz, S. et al. Exploring whole proteome to contrive multi-epitope-based vaccine for NeoCoV: An immunoinformtics andin-silicoapproach. Front. Immunol. 13, 956776. https://doi.org/10.3389/fimmu.2022.956776 (2022).
Cheng, P. et al. Evaluation of the consistence between the results of immunoinformatics predictions and real-world animal experiments of a new tuberculosis vaccine MP3RT. Front. Cell. Infect. Microbiol. 12, 1047306. https://doi.org/10.3389/fcimb.2022.1047306 (2022).
Pan, J. et al. An intranasal multivalent epitope-based nanoparticle vaccine confers broad protection against divergent influenza viruses. ACS Nano 17(14), 13474–13487. https://doi.org/10.1021/acsnano.3c01829 (2023).
Funding
This research was supported by the Science and Technology Program of Guangdong Province (2021B1212030015), Shihezi University Youth Innovation Talent Program Project (CXBJ202202), National Natural Science Foundation of China (32060811).
Author information
Authors and Affiliations
Contributions
S.L. (Shinian Li) and Y.L. (Yaling Li) came up with and designed the study; Formal analysis J.Y. (Jingjing Yu); writing—original draft, S.L. and C.X. (Chencheng Xiao); writing—review and editing, Y.L and C.X.; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, S., Yu, J., Xiao, C. et al. Immunoinformatics method to design universal multi-epitope nanoparticle vaccine for TGEV S protein. Sci Rep 15, 10931 (2025). https://doi.org/10.1038/s41598-025-95602-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-95602-w