Abstract
Acid Magenta, also known as Fuchsine acid, is a polycrystalline material with an anorthic structure that stands out for its eco-friendliness, cost-effectiveness, and versatility. Widely used in histology for cellular differentiation and across various industrial applications, Acid Magenta’s detailed molecular understanding and prediction of physicochemical properties hold significant value. Despite its relevance, research focused on the molecular topological modeling of Acid Magenta remains limited. This gap presents a unique opportunity to develop accurate and reliable topological descriptions of this molecular structure, which could significantly benefit industrial applications. This study aims to bridge the existing gap in topological modeling of Acid Magenta through a Quantitative Structure–Property Relationship (QSPR) and Quantitative Structure–Activity Relationship (QSAR) analysis, potentially enhancing its industrial utility. Utilizing efficient vertex partitioning techniques from chemical graph theory, we developed closed-form expressions for crucial topological descriptors, including the 1st multiplicative Zagreb index, 1st exponential Zagreb index, forgotten topological index, inverse degree index, 1st Zagreb index, multiplicative exponential Zagreb index and modified 1st Zagreb index. These derived indices were systematically evaluated to assess their impact on the structural properties of Acid Magenta. To further test predictive potential, the model applied linear fitting to key physicochemical properties of 18 octane isomers. Findings reveal that the indices 1st Zagreb index, Forgotten index, and Inverse degree index exhibit strong predictive correlations with these properties, particularly for entropy, the acentric factor, and enthalpy of vaporization (HVAP). The robust predictive performance of these indices underscores their potential as efficient alternatives to experimental testing for future QSAR/QSPR analysis, paving the way for new applications in environmental and industrial settings.
Similar content being viewed by others
Introduction
In recent years, organic semiconductors have garnered significant attention in the fields of electronics and photonic devices due to their versatile applications in optoelectronics, space technology, sensor fabrication, solar cells, and memory devices1. These materials are particularly advantageous because their π-electron systems allow them to transition into a conducting state, which is highly dependent on their unique chemical structures1,2. Among these, Acid Magenta stands out as a promising organic semiconductor. Naturally, it exhibits a polycrystalline structure with an anorthic type lattice, making it both environmentally friendly and cost-effective. Additionally, its ease of synthesis and handling enhances its appeal for various applications.
Fuchsine (\(\:{C}_{20}{H}_{19}{N}_{3}HCl\left[m,\:n\right]\)) is a triamine salt with two main amines and a secondary amine. A blend of rosaniline, pararosaniline, new fuchsine, and magenta II makes up basic fuchsine. Acid Magenta (C20H17N3Na2O9S3[m, n]) is a combination of homologues of basic fuchsine that have had sulfonic groups added to them. This modification not only imparts the dye with a distinctive greenish-yellow hue and high metallic luster but also makes it exceptionally soft, comparable to or softer than talc. Acid Magenta is widely used as a histological dye to enhance contrast in microscopic and electron microscopic examinations, helping to visualize specific cellular components, such as nuclei and cytoplasm. Its ability to differentiate cells from surrounding connective tissues further underscores its utility in biological and medical research.
In the context of chemical graph theory, the molecular structure of compounds like Acid Magenta can be represented numerically using topological indices, which capture the underlying topology of a molecule’s graph. These indices are crucial in establishing correlations between molecular structures and their physico-chemical properties, playing an essential role in quantitative structure-activity relationships (QSAR) and quantitative structure-property relationships (QSPR). Among the earliest and most widely studied topological descriptors are the Zagreb indices, originally introduced to estimate the total π-electron energy of molecules. These indices are calculated from the degrees of vertices within the molecular graph, offering insights into the electronic structure and reactivity of the compounds. It was demonstrated that the total π-electron energy \(\:\left(E\right)\) of a molecule \(\:M\) depends on the quantity \(\:\:\sum\:_{y\in\:V\left(G\right)}{\delta\:\left(y\right)}^{2}\) (now a days called the first Zagreb index), where \(\:G\) is the graph of the molecule \(\:M,\:\) \(\:V\left(G\right)\) corresponds to the vertex set of \(\:G\) and \(\:\delta\:\left(y\right)\) corresponds to degree of the vertex \(\:y\) (number of vertices adjacent to a vertex \(\:y\) in a graph \(\:G\) is equal to its degree). In molecular graph theory, chemical structure of a chemical compound /molecule is represented by its molecular graph. The molecular structure descriptor correlates with many physico-chemical characteristics of the concerned chemical compounds. These descriptors of molecular structure are commonly called as topological indices which are numeric parameters derived from molecular graphs that are invariant under automorphism. Thus, topological indices are an important tools for distinguishing isomers, and they have also demonstrated their utility in QSPR and QSAR3, and nanotechnology including drug discovery and design4. In mathematical chemistry, topological indices have various categories; distance-based indices5,6,7,8,9, spectrum-based indices10,11,12,13,14, vertex-degree-based indices15,16,17,18,19,20,21 and connection-based indices22,23,24,25,26.
In 1947, Wiener introduced the concept of a topological index during his studies on the boiling points of paraffins, coining it the Wiener index27. This foundational work laid the groundwork for subsequent research in molecular graph theory. Building on Wiener’s contributions, M. Darafsheh28 developed methods to calculate both the Wiener and Szeged indices across various graph structures. In addition, A. Ayache29 extended these concepts by calculating topological indices specifically for mk-graphs, while Wei Gao et al.30 explored eccentricity-based topological indices within the cycloalkane family. Topological indices that are based on vertex degrees have proven particularly effective for correlating molecular structures with their physico-chemical properties. These indices are essential in establishing quantitative relationships between structure and behavior in chemical compounds. Ullah and Zaman have made significant contributions in this area, conducting extensive research on degree-based topological descriptors for a wide range of molecular structures31,32,33,34,35,36,37,38,39,40,41,42,43,44. For additional insights into the topological characterization of molecular and microstructures, the refs45,46,47,48,49,50,51,52,53,54. offer a comprehensive exploration of this subject.
The 1st Zagreb index55 is the oldest vertex degree based index, defined as:
This index, along with its extensions such as the multiplicative Zagreb indices, exponential Zagreb indices, and other vertex-degree-based descriptors, has been extensively studied for its utility in understanding molecular properties. Notably, these indices are invaluable for distinguishing isomers and optimizing molecular structures for applications in fields such as drug discovery, nanotechnology, and materials science. For detailed understanding of these indices, reader can consult refs56. and citations therein. Though in the literature, many types of Zagreb indices are introduced but the 1st and 2nd multiplicative Zagreb indices57, Zagreb Co-indices58, and59 are most valued. The multiplicative exponential Zagreb indices, the 1st exponential Zagreb index, and 2nd exponential Zagreb index for \(\:{C}_{n}\:,\:\:{K}_{n}\:,\:{P}_{n}\:,\:{S}_{n}\:,\:{W}_{n}\:\)are studied in60. Notations and terminology can be found in refs45,46,61. except if otherwise stated, are not defined here.
Consider G is a molecular graph, \(\:V\left(G\right)\) and \(\:E\left(G\right)\) are vertex and edge sets of a graph \(\:G\). The degree of vertex in any graph G refers to number of edges connected to a given vertex. It is represented by \(\:\delta\:\left(y\right)\).
Gutman and Trinajstić introduced the 1st Zagreb index over 30 years ago55, and is defined as
The 1st Multiplicative Zagreb Index, presented by Todeschini et al.62, can be defined as
The modified 1st Zagreb index was proposed in63, it is defined as
The forgotten topological index64 can be defined as
The inverse degree index65, a vertex based graph invariant is defined as
The first exponential Zagreb index60, introduced by Nihat Akgunes and Busra Aydin, is define as
Multiplicative exponential Zagreb index60 can be defined as
This study is driven by the substantial industrial significance and diverse applications of Acid Magenta, particularly in dye manufacturing, histology, and materials science. Our primary objective is to leverage chemical graph theory to model the molecular structure of Acid Magenta using vertex partitioning techniques. By doing so, we aim to compute a series of topological descriptors, including those mentioned above (Eqs. 1–7), and derive closed-form expressions for them. This mathematical representation will provide a deeper understanding of the structural properties of Acid Magenta, enhancing its potential for applications in QSAR/QSPR analysis, thus paving the way for future innovations in the design of organic semiconductors and related materials.
Molecular topological modeling of acid magenta
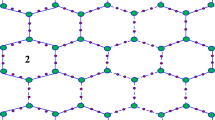
The 2D graph of Acid Magenta molecular structure is given in Fig. 1. We define \(\:m\) as the number of linked unit cells in a row and \(\:n\) as the number of associated rows with each number of \(\:m\) cells. The order of the following molecular graph is \(\:38mn+m+n,\:\)and total number of edges are \(\:42mn\). To abstract the topological indices mentioned in Sect. 1, we have partitioned the total number of vertices in three different sets, say \(\:{V}_{1}\), \(\:{V}_{2}\:,\) and \(\:{V}_{3}\). The set \(\:{V}_{1}\) contains those vertices which have degree\(\:\:1\). Set \(\:{V}_{2}\) contain the vertices with degree two and set \(\:{V}_{3}\) contain the vertices with degree three, respectively. Table 1 shows vertex partition of Acid Magenta’s graph.
Chemical structure of Acid Magenta (a) 3D (b) 2D (c) Graph theoretic model of the chemical structure of Acid Magenta, C20H17N3Na2O9S31.
Main results
Derivation of closed formulae for the topological indices of acid magenta
Consider the graph \(\:G\cong\:\:\) C20H17N3Na2O9S3[m, n] of molecular structure of Acid Magenta with \(\:m,\:n\ge\:1,\:\)then;
-
(1)
\(M_{1} \left( G \right) = ~220mn - 2m - 2n\)
-
(2)
\(\mathop \prod \limits_{1} \left( G \right)~ = ~~~1584mn\left( { - m + 2mn - n} \right)\left( {m + 7mn + n} \right)\)
-
(3)
\(M_{1}^{*} \left( G \right) = ~\frac{{305mn}}{{18}} + \frac{{7m}}{4} + \frac{{7n}}{4}\)
-
(4)
\(F_{N} \left( G \right) = ~624mn - 6m - 6n\)
-
(5)
\(ID\left( G \right) = ~\frac{1}{6}\left( {9n + m\left( {9 + 134n} \right)} \right)\)
-
(6)
\(EM_{1} \left( G \right) = 178415.096mn - 49.162m - 49.162n\)
-
(7)
\(E\mathop \prod \limits_{1} \left( G \right)~ = 52909000mn\left( {2mn - m - n} \right)\left( {7mn + m + n} \right)\)
Proofs
Consider \(\:G\:\)as graph of Acid Magenta. By the help of Table 1 and Eqs. 1–7 we will compute the following TIs and will derive formulas for them.
-
(1)
The first Zagreb index
By Table 1 and Eq. (1), value of 1st Zagreb index is;
$$M_{1} \left( G \right) = \sum\limits_{{y \in V\left( G \right)}} {\delta \left( y \right)} ~^{2}$$$$\begin{aligned} M_{1} \left( G \right) = & ~\mathop \sum \limits_{{y \in V_{1} \left( G \right)}} \delta \left( y \right)^{2} + ~\mathop \sum \limits_{{y \in V_{2} \left( G \right)}} \delta \left( y \right)^{2} + ~\mathop \sum \limits_{{y \in V_{3} \left( G \right)}} \delta \left( y \right)^{2} \\ ~ = & ~~\left( 1 \right)^{2} \left( {14mn + 2\left( {m + n} \right)} \right) + \left( 2 \right)^{2} \left( {2mn - m - n} \right) + ~\left( 3 \right)^{2} \left( {22mn} \right) \\ ~ = & ~~220mn - 2m - 2n \\ \end{aligned}$$ -
(2)
The first multiplicative Zagreb index
By Table 1 and Eq. (2), the value of 1st multiplicative Zagreb index is;
$$\:{\prod\:}_{1}\left(G\right)=\:\prod\:_{y\in\:V\left(G\right)}{\delta\:\left(y\right)}^{2}$$$$\begin{aligned} \mathop \prod \limits_{1} \left( G \right) = ~ & \mathop \prod \limits_{{y \in V_{3} \left( G \right)}} \delta \left( y \right)^{2} \times ~~\mathop \prod \limits_{{y \in V_{2} \left( G \right)}} \delta \left( y \right)^{2} \times ~\mathop \prod \limits_{{y \in V_{3} \left( G \right)}} \delta \left( y \right)^{2} \\ = & \left( 1 \right)^{2} \left( {14mn + 2\left( {m + n} \right)} \right) \times \left( 2 \right)^{2} \left( {2mn - m - n} \right) \times ~\left( 3 \right)^{2} \left( {22mn} \right) \\ ~ = & ~~1584mn\left( { - m + 2mn - n} \right)\left( {m + 7mn + n} \right) \\ \end{aligned}$$ -
(3)
The modified first Zagreb index
We compute the modified 1st Zagreb index by Table 1 and Eq. (3) as;
$$\:{M}_{1}^{*}\left(G\right)=\:\sum\:_{y\in\:V\left(G\right)}\frac{1}{{\delta\:\left(y\right)}^{2}}$$$$\begin{aligned} M_{1}^{*} \left( G \right) = & ~\mathop \sum \limits_{{y \in V\left( G \right)}} \frac{1}{{\delta \left( y \right)^{2} }} + \mathop \sum \limits_{{y \in V\left( G \right)}} \frac{1}{{\delta \left( y \right)^{2} }} + \mathop \sum \limits_{{y \in V\left( G \right)}} \frac{1}{{\delta \left( y \right)^{2} }} \\ ~ = & \left( {14mn + 2m + 2n} \right) + \frac{1}{4}\left( {2mn - m - n} \right) + \frac{1}{9}\left( {22mn} \right) \\ = & ~\frac{{305mn}}{{18}} + \frac{{7m}}{4} + \frac{{7n}}{4} \\ \end{aligned}$$ -
(4)
The forgotten topological index
The value of forgotten topological index is computed using Table 1 and Eq. (4) as follow
$$\:{F}_{N}\left(G\right)=\:\sum\:_{y\in\:V\left(G\right)}{\delta\:\left(y\right)}^{3}$$$$\begin{aligned} F_{N} \left( G \right) = ~ & \mathop \sum \limits_{{y \in V_{1} \left( G \right)}} \delta \left( y \right)^{3} + ~\mathop \sum \limits_{{y \in V_{2} \left( G \right)}} \delta \left( y \right)^{3} + ~\mathop \sum \limits_{{y \in V_{3} \left( G \right)}} \delta \left( y \right)^{3} \\ ~ = & ~~\left( 1 \right)^{3} \left( {14mn + 2\left( {m + n} \right)} \right) + \left( 2 \right)^{3} \left( {2mn - m - n} \right) + ~\left( 3 \right)^{3} \left( {22mn} \right) \\ ~ = ~ & ~624mn - 6m - 6n \\ \end{aligned}$$ -
(5)
The inverse degree index
We compute the inverse degree index by the help of Table 1 and Eq. (5) as;
$$\:ID\left(G\right)=\sum\:_{y\in\:V\left(G\right)}\frac{1}{\delta\:\left(y\right)}$$$$\begin{aligned} ID\left( G \right) = & \mathop \sum \limits_{{y \in V_{1} \left( G \right)}} \frac{1}{{\delta \left( y \right)}} + \mathop \sum \limits_{{y \in V_{2} \left( G \right)}} \frac{1}{{\delta \left( y \right)}} + \mathop \sum \limits_{{y \in V_{3} \left( G \right)}} \frac{1}{{\delta \left( y \right)}} \\ = & ~\left( {14mn + 2m + 2n} \right) + \frac{1}{2}\left( {2mn - m - n} \right) + \frac{1}{3}\left( {22mn} \right) \\ = & ~\frac{1}{6}\left( {9n + m\left( {9 + 134n} \right)} \right) \\ \end{aligned}$$ -
(6)
The first exponential Zagreb index
We compute 1st exponential Zagreb index using Table 1 and Eq. (6) as;
$$\:{EM}_{1}\left(G\right)=\sum\:_{y\in\:V\left(G\right)}{e}^{{\delta\:\left(y\right)}^{2}}$$$$\begin{aligned} EM_{1} \left( G \right) = & \mathop \sum \limits_{{y \in V_{1} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} + \mathop \sum \limits_{{y \in V_{2} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} + \mathop \sum \limits_{{y \in V_{3} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} \\ ~ = ~ & ~e^{{\left( 1 \right)^{2} }} \left( {14mn + 2m + 2n} \right) + e^{{\left( 2 \right)^{2} }} \left( {2mn - m - n} \right) + e^{{\left( 3 \right)^{2} }} \left( {22mn} \right) \\ ~ = & ~~178415.096mn - 49.162m - 49.162n \\ \end{aligned}$$ -
(7)
The multiplicative exponential Zagreb index
We compute the multiplicative exponential Zagreb index by using Table 1 and Eq. (7) as;
$$\:E{\prod\:}_{1}\left(G\right)=\:\prod\:_{y\in\:V\left(G\right)}{e}^{{\delta\:\left(y\right)}^{2}}$$$$\begin{aligned} E\mathop \prod \limits_{1} \left( G \right)~ = ~ & \mathop \prod \limits_{{y \in V_{1} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} ~ \times ~\mathop \prod \limits_{{y \in V_{2} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} \times ~\mathop \prod \limits_{{y \in V_{3} \left( G \right)}} e^{{\delta \left( y \right)^{2} }} \\ = ~ & ~e^{{\left( 1 \right)^{2} }} \left( {14mn + 2m + 2n} \right) \times e^{{\left( 2 \right)^{2} }} \left( {2mn - m - n} \right) \times e^{{\left( 3 \right)^{2} }} \left( {22mn} \right) \\ = & 52909000mn\left( {2mn - m - n} \right)\left( {7mn + m + n} \right) \\ \end{aligned}$$
Numerical analysis and graphical visualization of computed topological indices
The applications of Degree-based topological indices are extensive across various fields, like pharmaceuticals, chemistry, biomedical sciences, and computer science. These indices provide scientists with valuable numerical and graphical insights into the analyzed data. In this study, we have numerically calculated various degree-based topological indices for Acid Magenta at diverse values of m and n. To facilitate comparison, we also compiled a table (Table 2) that presents these indices for varying m and n values, enabling a straightforward numerical comparison of the molecular structure. As observed in Table 2, the indices consistently increase in value as the parameters m and n grow.
The graphical representation and comparison of our calculated results for varying values of m, and n in C20H17N3Na2O9S3 [m, n] can be seen in the Figs. 2, 3 and 4. It is evident from the Table 2; Figs. 2, 3 and 4, values of all given indices are increasing as we increase the corresponding values of m, n.
Testing the property prediction potential and effectiveness of the considered topological indices in QSPR analysis
Topological indices considered here can be used to predict some physicochemical properties and various substance parameters, including the acentric factor, B.P (boiling point), enthalpy of vaporization and entropy. According to guidelines of ‘The International Academy of Mathematical Chemistry (IAMC)’ to assess the effectiveness of any topological index in modeling physicochemical attributes regression analysis is commonly employed66. Octane isomers are particularly useful due to their structural diversity, which results in branching patterns, non-polar properties and forms. These organic compounds are ideal for statistical testing due their ample number of structural isomers and the availability of comprehensive experimental data for each of them. As noted by Randić and Trinajstić67, linking theoretical invariants to experimentally found physicochemical properties of octane isomers is beneficial for determining the predictive power of an invariant68,69.
In this study, firstly we computed topological indices for eighteen octane isomers and investigated the correlation of the calculated values and experimental parameters to evaluate the predictive potential and practical applicability of these indices. The experimental values were sourced from PubChem, Chemspider databases and study66. The calculation of topological indices for these isomers were carried out by following methods similar to those described in Sect. 3.1.
Through linear fitting, we investigated four physicochemical characteristics of eighteen octane isomers. The analysis and visualization of results were conducted using Origin, MATLAB, and Excel softwares. The correlations of the physicochemical characteristics of octane isomers and their topological indices are illustrated in Figs. 5, 6, 7, 8, 9 and 10. The values of correlation coefficients are presented in Table 3.
It can be seen from Figs. 5, 6, 7, 8, 9 and 10; Table 3 that M1(G), FN(G) and ID(G) have strong correlations with all the four physical properties, M1*(G) has good correlation value for HVAP (Enthalpy of Vaporization) and EM1(G) has good correlation for DHVAP (Standard Enthalpy of Vaporization) and HVAP. It can also be seen that, among all these indices, ID(G) is the best predictor of entropy, Acentric Factor and DHVAP, and M1(G) is the best predictor of HVAP having strongest correlation. However, ∏_1(G) index does not show good correlation to model and predict any property. Here, the weaker associations of some indices with certain properties indicate that they are influenced by factors beyond the scope of these specific topological indices alone, such as intermolecular interactions or external conditions that are not fully encapsulated by concerned indices.
Conclusion
Molecular descriptors offer crucial insights into the chemical structure of molecules, playing a significant role in analyzing the QSPR (Quantitative Structure–Property Relationships) and QSAR (Quantitative Structure–Activity Relationships). These models are essential for correlating molecular structures or properties with descriptors that can predict chemical and biological activities. In this study, we focused on modeling and topologically characterizing the molecular structure of Acid Magenta using vertex partitioning techniques. We successfully computed and derived various expressions for several key topological indices; the 1st Zagreb index, 1st Multiplicative Zagreb index, Forgotten Topological index, Modified 1st Zagreb index, Inverse Degree index, 1st Exponential Zagreb index, and Multiplicative Exponential Zagreb index. The numerical results obtained from these models, across various values of m and n, demonstrated the effectiveness of all these topological indices to predict the physicochemical properties. To test the predictive potential of the above mentioned indices, we investigated four physicochemical characteristics of all the eighteen octane isomers through linear fitting. The results show that, the indices M1(G), FN(G) and ID(G) have strong correlations with all the four physical properties, M1*(G) has good correlation with HVAP and EM1(G) has good correlation with HVAP and DHVAP. Among all these indices, ID(G) is the best predictor of entropy, Acentric Factor and DHVAP, and M1(G) is the best predictor of HVAP having strongest correlation. These findings underscore the importance of the derived formulas in accurately modeling the molecular structure of Acid Magenta, providing valuable tools for future QSAR/QSPR analysis.
Data availability
All data generated or analyzed during this study are included within this article.
References
El-Zaidia, E. F. M., Al-Kotb, M. S. & Yahia, I. S. Physico-chemical properties of acid Fuchsin as novel organic semiconductors: Structure, optical and electrical properties. Phys. B: Condens. Matter 571, 71–75 (2019).
Kazheva, O. N., Kushch, N. D., Aleksandrov, G. G. & Dyachenko, O. A. Crystal structure and molecular packing in a new organic semiconductor penta[bis(ethylenedithio)tetrathiafulvalene]-hexathiocyanatomononitratoyttrium(III)-ethanol. Mater. Chem. Phys. 73, 193–197 (2002).
Zhang, Q. et al. Mathematical study of silicate and oxide networks through Revan topological descriptors for exploring molecular complexity and connectivity. Sci. Rep. 15, 8116 (2025)
Khadikar, P. V. Padmakar-Ivan Index in Nanotechnology (IRANIAN JOURNAL OF MATHEMATICAL CHEMISTRYs, 2010).
Hayat, S. Distance-based graphical indices for predicting thermodynamic properties of benzenoid hydrocarbons with applications. Comput. Mater. Sci. 230, 112492 (2023).
Liu, J. B., Gu, J. J. & Wang, K. The expected values for the Gutman index, Schultz index, and some Sombor indices of a random cyclooctane chain. Int. J. Quantum Chem. 123, e27022 (2022).
Arockiaraj, M., Klavzar, S., Clement, J., Mushtaq, S. & Balasubramanian, K. Edge Distance-based topological indices of Strength-weighted graphs and their application to coronoid systems, carbon nanocones and SiO(2) nanostructures. Mol. Inf. 38, e1900039 (2019).
Zaman, S., Kamboh, A., Ullah, A. & Liu, J. B. Development of some novel resistance distance based topological indices for certain special types of graph networks. Phys. Scr. 98, 125250 (2023).
Ullah, A., Salman, S. & Zaman, S. Resistance distance and Sharp bounds of two-mode electrical networks. Phys. Scr. 99, 085241 (2024).
Zaman, S. & Ullah, A. Kemeny’s constant and global mean first passage time of random walks on octagonal cell network. Math. Methods Appl. Sci. 46, 9177–9186 (2023).
Zaman, S., Mustafa, M., Ullah, A. & Siddiqui, M. K. Study of mean-first-passage time and Kemeny’s constant of a random walk by normalized laplacian matrices of a penta-chain network. Eur. Phys. J. Plus 138, 770 (2023).
Yu, X., Zaman, S., Ullah, A., Saeedi, G. & Zhang, X. Matrix analysis of hexagonal model and its applications in global Mean-First-Passage time of random walks. IEEE Access. 11, 10045–10052 (2023).
Yan, T., Kosar, Z., Aslam, A., Zaman, S. & Ullah, A. Spectral techniques and mathematical aspects of K4 chain graph. Phys. Scr. 98, 045222 (2023).
Kosar, Z., Zaman, S., Ali, W. & Ullah, A. The number of spanning trees in a K5 chain graph. Phys. Scr., DOI (2023).
Hayat, S., Khan, M. A., Khan, A., Jamil, H. & Malik, M. Y. H. Extremal hyper-Zagreb index of trees of given segments with applications to regression modeling in QSPR studies. Alexandria Eng. J. 80, 259–268 (2023).
Hayat, S. & Asmat, F. Sharp Bounds on the Generalized Multiplicative First Zagreb Index of Graphs with Application to QSPR Modeling, Mathematics, (2023).
Liu, J. B., Ali, H., Shafiq, M. K., Dustigeer, G. & Ali, P. On topological properties of planar octahedron networks. Polycycl. Aromat. Compd. 43, 755–771 (2023).
Raza, Z., Arockiaraj, M., Maaran, A., Kavitha, S. R. J. & Balasubramanian, K. Topological entropy characterization, NMR and ESR spectral patterns of Coronene-Based transition metal organic frameworks. ACS Omega. 8, 13371–13383 (2023).
Arockiaraj, M. et al. Novel molecular hybrid geometric-harmonic-Zagreb degree based descriptors and their efficacy in QSPR studies of polycyclic aromatic hydrocarbons. SAR QSAR Environ. Res. 34, 569–589 (2023).
Mondal, S., De, N. & Pal, A. Topological indices of some chemical structures applied for the treatment of COVID-19 patients. J. Polycycl. Aromatic Compd. 42, 1220–1234 (2022).
Kirana, B., Shanmukha, M. C. & Usha, A. Comparative study of Sombor index and its various versions using regression models for top priority polycyclic aromatic hydrocarbons. Sci. Rep. 14, 19841 (2024).
Usman, M. & Javaid, M. Connection-Based Zagreb indices of polycyclic aromatic hydrocarbons structures, curr org synth, (2023). https://doi.org/10.2174/1570179421666230823141758
Das, K. C., Mondal, S. & Raza, Z. On Zagreb connection indices. Eur. Phys. J. Plus. 137, 1242 (2022).
Ullah, A., Jamal, M., Zaman, S. & Shamsudin Connection based novel AL topological descriptors and structural property of the zinc oxide metal organic frameworks. Phys. Scr. 99, 055202 (2024).
Li, X. et al. Computational insights into zinc silicate MOF structures: topological modeling, structural characterization and chemical predictions. Sci. Rep. 14, 19866 (2024).
Ullah, A., Udin, S., Zaman, S. & Hamraz, A. Zagreb connection topological descriptors and structural property of the triangular chain structures. Phys. Scr. 98, 025009 (2023).
Wiener, H. Structural determination of paraffin boiling points. J. Am. Chem. Soc. 69, 17–20 (1947).
Darafsheh, M. R. Computation of topological indices of some graphs. Acta Applicandae Math. 110, 1225–1235 (2010).
Ayache, A. & Alameri, A. Topological indices of the mk-graph. J. Association Arab. Universities Basic. Appl. Sci. 24, 283–291 (2017).
Gao, W., Chen, Y. & Wang, W. The topological variable computation for a special type of cycloalkanes. J. Chem. 2017, 6534758 (2017).
Ullah, A., Jabeen, S., Zaman, S., Hamraz, A. & Meherban, S. Predictive potential of K-Banhatti and Zagreb type molecular descriptors in structure–property relationship analysis of some novel drug molecules. J. Chin. Chem. Soc. 71, 250–276 (2024).
Ullah, A., Bano, Z. & Zaman, S. Computational aspects of two important biochemical networks with respect to some novel molecular descriptors. J. Biomol. Struct. Dynamics. 42, 791–805 (2024).
Zaman, S., Yaqoob, H. S. A., Ullah, A. & Sheikh, M. QSPR analysis of some novel drugs used in blood cancer treatment via degree based topological indices and regression models, polycyclic aromatic compounds, (2023). https://doi.org/10.1080/10406638.2023.2217990 1–17 .
Zaman, S., Ullah, A. & Shafaqat, A. Structural modeling and topological characterization of three kinds of dendrimer networks. Eur. Phys. J. E Soft Matter. 46, 36 (2023).
Zaman, S. et al. Three-Dimensional Structural Modelling and Characterization of Sodalite Material Network concerning the Irregularity Topological Indices, J. Math. (2023) 1–9. (2023).
Zaman, S., Jalani, M., Ullah, A., Ali, M. & Shahzadi, T. On the topological descriptors and structural analysis of cerium oxide nanostructures. Chem. Pap. 77, 2917–2922 (2023).
Zaman, S., Jalani, M., Ullah, A., Ahmad, W. & Saeedi, G. Mathematical analysis and molecular descriptors of two novel metal–organic models with chemical applications. Sci. Rep. 13, 5314 (2023).
Ullah, A., Zaman, S., Hussain, A., Jabeen, A. & Belay, M. B. Derivation of mathematical closed form expressions for certain irregular topological indices of 2D nanotubes. Sci. Rep. 13, 11187 (2023).
Ullah, A., Zaman, S., Hamraz, A. & Muzammal, M. On the construction of some bioconjugate networks and their structural modeling via irregularity topological indices. Eur. Phys. J. E 46, 72 (2023).
Ullah, A., Shamsudin, S., Zaman, A. & Hamraz Zagreb connection topological descriptors and structural property of the triangular chain structures. Phys. Scr. 98, 025009 (2023).
Hakeem, A., Ullah, A. & Zaman, S. Computation of some important degree-based topological indices for γ- Graphyne and zigzag Graphyne nanoribbon. Mol. Phys. 121, e2211403 (2023).
Zaman, S., Jalani, M., Ullah, A., Saeedi, G. & Guardo, E. Structural Analysis and Topological Characterization of Sudoku Nanosheet, Journal of Mathematics, (2022) 1–10. (2022).
Ullah, A. et al. Network-Based Modeling of the Molecular Topology of Fuchsine Acid Dye with Respect to Some Irregular Molecular Descriptors, Journal of Chemistry, (2022) 1–8. (2022).
A, S. M. C. U. & B. N S, S. K. C. Comparative study of multilayered graphene using numerical descriptors through M-polynomial. Phys. Scr. 98, 075205 (2023).
Ullah, A., Zeb, A. & Zaman, S. A new perspective on the modeling and topological characterization of H-Naphtalenic nanosheets with applications. J. Mol. Model. 28, 211 (2022).
Ullah, A., Qasim, M., Zaman, S. & Khan, A. Computational and comparative aspects of two carbon nanosheets with respect to some novel topological indices. Ain Shams Eng. J. 13, 101672 (2022).
Zhang, X., Aslam, A., Saeed, S., Razzaque, A. & Kanwal, S. Investigation for metallic crystals through chemical invariants, QSPR and fuzzy-TOPSIS. J. Biomol. Struct. Dyn. 1–12. https://doi.org/10.1080/07391102.2023.2209656 (2023).
Iqbal, Z., Ishaq, M., Aslam, A., Aamir, M. & Gao, W. The measure of irregularities of nanosheets. Open. Phys. 18, 419–431 (2020).
Khabyah, A. A., Zaman, S., Koam, A. N. A., Ahmad, A. & Ullah, A. minimum zagreb eccentricity indices of two-mode network with applications in boiling point and benzenoid hydrocarbons, Mathematics, 10 (2022).
Ullah, A., Shaheen, M., Khan, A., Khan, M. & Iqbal, K. Evaluation of topology-dependent growth rate equations of three-dimensional grains using realistic microstructure simulations. Mater. Res. Express 6, 026523 (2018).
Ullah, A. et al. Simulations of grain growth in realistic 3D polycrystalline microstructures and the MacPherson–Srolovitz equation. Mater. Res. Express 4, 066502 (2017).
Asad, U. et al. Neighborhood topological effect on grain topology-size relationship in three-dimensional polycrystalline microstructures. Chin. Sci. Bull. 58, 3704–3708 (2013).
Kirana, B., Shanmukha, M. C. & Usha, A. A QSPR Analysis and Curvilinear Regression Models for Various degree-based Topological Indices: Quinolone Antibiotics 10 (Heliyon, 2024).
Shanmukha, M. C. et al. Chemical applicability and computation of K-Banhatti indices for benzenoid hydrocarbons and triazine-based covalent organic frameworks. Sci. Rep. 13, 17743 (2023).
Gutman, I. & Trinajstić, N. Graph theory and molecular orbitals. Total φ-electron energy of alternant hydrocarbons. Chem. Phys. Lett. 17, 535–538 (1972).
Das, K. C., Yurttas, A., Togan, M. & Cevik, A. S. C. Ismail Naci, The multiplicative Zagreb indices of graph operations, Journal of inequalities applications, (2013) 1–14. (2013).
Eliasi, M., Iranmanesh, A. & Gutman, I. Multiplicative versions of first Zagreb index. Match-Communications Math. Comput. Chem. 68, 217 (2012).
Das, K. C. et al. On the first Zagreb index and multiplicative Zagreb coindices of graphs, analele universitatii Ovidius. Constanta-Seria Matematica. 24, 153–176 (2016).
Hua, H., R Ashrafi, A. & Zhang, L. More on Zagreb coindices of graphs. Filomat 26, 1215–1225 (2012).
Akgunes, N. & Aydin, B. Introducing New Exponential Zagreb Indices for Graphs, Journal of Mathematics, (2021). (2021).
Shanmukha, M. C., Gowtham, K. J., Usha, A. & Julietraja, K. Expected values of Sombor indices and their entropy measures for graphene. Mol. Phys. 122, e2276905 (2024).
Todeschini, R., Ballabio, D. & Consonni, V. Novel molecular descriptors based on functions of new vertex degrees, DOI (2010).
Rodríguez, J. M., Sánchez, J. L. & Sigarreta, J. M. Inequalities on the inverse degree index. J. Math. Chem. 57, 1524–1542 (2019).
Furtula, B. & Gutman, I. A forgotten topological index. J. Math. Chem. 53, 1184–1190 (2015).
Fajtlowicz, S. On conjectures of Graffiti-II. Congr Numer. 60, 187–197 (1987).
Mondal, S., De, N. & Pal, A. On neighborhood Zagreb index of product graphs. J. Mol. Struct. 1223, 129210 (2021).
Randić, M. & Trinajstić, N. In search for graph invariants of chemical interes. J. Mol. Struct. 300, 551–571 (1993).
Mondal, S., Dey, A., De, N. & Pal, A. QSPR analysis of some novel neighbourhood degree-based topological descriptors. Complex. Intell. Syst. 7, 977–996 (2021).
Randic, M., Guo, X., Oxley, T., Krishnapriyan, H. & Naylor, L. Wiener matrix invariants. J. Chem. Inf. Comput. Sci. 34, 361–367 (2002).
Acknowledgements
The authors extend their appreciation to Taif University, Saudi Arabia, for supporting this work through project number (TU-DSPP-2024-47). The authors also express their gratitude for the financial support from Natural Science Fund project of Universities in Anhui Province (No. 2024AH050616, 2022AH052889) and Anhui Xinhua University Scientific Research Project (2024zr009).
Funding
This research was funded by Taif University, Saudi Arabia, Project No. (TU-DSPP-2024-47), Natural Science Fund project of Universities in Anhui Province (No. 2024AH050616, 2022AH052889) and Anhui Xinhua University Scientific Research Project (2024zr009).
Author information
Authors and Affiliations
Contributions
All the authors Jiang-Hua Tang, Anila Hamraz, Asad Ullah, Y. S. Hamed, Shahid Zaman, and Melaku Berhe Belay have equally contributed to this manuscript in all stages from conceptualization to the write up of final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All authors have approved the manuscript and given consent for publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, JH., Hamraz, A., Ullah, A. et al. Chemical applicability and predictive potential of certain graphical indices for determining structure-property relationships in polycrystalline acid magenta (C20H17N3Na2O9S3). Sci Rep 15, 13886 (2025). https://doi.org/10.1038/s41598-025-98024-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-98024-w
Keywords
This article is cited by
-
Topological analysis of the structural complexity and irregularity of Kudriavite (CdBi2S4)
Chemical Papers (2025)