Abstract
Osteomyelitis, characterized by bone inflammation and infection, poses a significant global health burden. This Mendelian randomization (MR) study investigates the causal relationship between polyunsaturated fatty acids (PUFAs) and osteomyelitis risk. By using GWAS data from 114,999 individuals, we explore specific PUFAs and their genetic variations using Inverse variance weighted (IVW), MR-Egger and weighted median methods. The results reveal a suggestive association between genetically predicted higher docosahexaenoic acid (DHA) and omega-6 levels with increased osteomyelitis risk. Conversely, a negative association is found for the omega-6:3 ratio. Linoleic acid, omega-3, and omega-6 show no significant associations. Heterogeneity and pleiotropy analyses support result robustness, indicating minimal confounding effects. Sensitivity analyses confirm the stability of findings. Our MR analysis challenges the presumed protective role of omega-3 in osteomyelitis, suggesting a nuanced relationship where DHA may pose an increased risk. The study underscores the complexity of fatty acid interactions influenced by genetic variability and dietary nuances. Further research is essential to unravel underlying mechanisms and translate these findings into actionable strategies for osteomyelitis prevention and treatment.
Similar content being viewed by others
Introduction
Osteomyelitis, a debilitating condition characterized by the inflammation and infection of bone tissue, poses a significant burden on individuals’ health and healthcare systems worldwide1,2,3. It results from bacterial or fungal pathogens infiltrating the bone, leading to severe pain, limited mobility, and, in some cases, life-threatening complications4,5. Osteomyelitis is often refractory to treatment, with its occurrence and severity influenced by a complex interplay of genetic predisposition and environmental factors6,7. Understanding the causative factors and risk determinants behind this condition is crucial for developing more effective preventive and therapeutic strategies, reducing the suffering it inflicts, and improving patient outcomes. Osteomyelitis results from microbial invasion of bone tissue, leading to inflammation and bone destruction. Systemic factors like nutritional status and immune function play critical roles in susceptibility and progression. Modulation of inflammatory responses and immune cell function, potentially influencing the body’s ability to combat bone infections.
Polyunsaturated fatty acids, including omega-3 and omega-6 fatty acids, have long been recognized for their potential health benefits8. These essential dietary components play critical roles in cellular function, inflammation regulation, and overall well-being9. Some studies have suggested that a higher intake of certain polyunsaturated fatty acids might confer protection against inflammatory and infectious diseases10,11,12,13,14,15. The combination of vancomycin with omega-3 fatty acids was effective against bacterial growth in animals with bone infection16. However, the exact nature of this relationship and its potential causal link with osteomyelitis has remained unclear. Understanding how polyunsaturated fatty acids interact with these health outcomes is of paramount importance for tailoring dietary recommendations and interventions that could positively impact public health.
Despite existing studies on PUFAs and osteomyelitis, there is a lack of evidence on the causal relationship due to confounding factors inherent in observational studies. Mendelian Randomization (MR) provides an innovative and rigorous framework for assessing causal relationships between exposures and disease outcomes17. By leveraging genetic variants as instrumental variables, MR can help disentangle the complexities of confounding factors and reverse causality that often plague observational studies18,19. To date, no studies have investigated the causal relationship between PUFAs and osteomyelitis using MR. The aim of this study is to employ MR to unravel the causal associations between polyunsaturated fatty acids and the risk of osteomyelitis. We hypothesize that higher genetically predicted levels of PUFAs, particularly omega-3 fatty acids, are associated with a decreased risk of osteomyelitis due to their anti-inflammatory properties.
Method
Study design
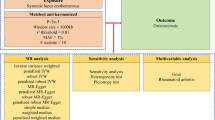
In our study, the summary statistics from genome-wide association studies (GWAS) were employed to investigate the potential link for osteomyelitis with various polyunsaturated fatty acids (PUFAs), specifically, docosahexaenoic acid (DHA), linoleic acid (LA), omega-3 fatty acids, omega-6 fatty acids, and the omega-6 to omega-3 ratio (omega-6) by MR approach. To ensure the validity of our instrumental variables, we confirmed that they satisfy the three core MR assumptions: relevance, independence, and exclusion restriction. The MR analysis was grounded in three essential principles to ensure its reliability and accuracy: firstly, the genetic variants selected needed to show a substantial association with the PUFAs under study; secondly, these variants should only affect the outcome (osteomyelitis) through their interaction with the PUFAs; and thirdly, there should be no links between these genetic variants and any confounding factors that could influence the PUFA-osteomyelitis relationship. The design of our study, which methodically addresses these key assumptions, is visually represented in Fig. 1. In alignment with the 2021 STROBE-MR guidelines, the findings of this study have been thoroughly reported, adhering to their recommended standards20.
Causal-directed acyclic graph. This figure was employed to illustrate the theoretical associations among a genetic variant denoted as G, an exposure marked as E, and an outcome represented as O. This graphical depiction accounts for the potential influence of unobserved confounding variables, labeled as C. In this diagram, solid arrows denote permissible relationships between these variables, reflecting known or hypothesized causal connections. Conversely, dashed lines are utilized to indicate relationships that are proscribed. These prohibited connections are crucial for G to meet the criteria for its validity as an instrumental variable (IV). It’s noteworthy that the arrows between G and E, as well as between E and O, are parameterized by 1 and 2, respectively. This parameterization serves to quantify the causal effect of E on O.
Genetic associations with PUFAs
The PUFAs as mentioned above were examined in the study. Genetic instrumentation for these PUFAs was obtained from 114,999 individuals, as recorded in the UK Biobank21. The UK Biobank, with its recruitment of over 500,000 Europeans, is aimed at uncovering the genetic and environmental factors that contribute to various diseases. The circulating levels of PUFAs were assessed using a targeted high-throughput nuclear magnetic resonance (NMR) metabolomics platform, referred to as the biomarker quantification version 2020. This platform’s measurement technology and its applications for epidemiological studies have been extensively evaluated in prior research22,23. Following the application of quality control criteria and the exclusion of duplicates from the initial collection of 121,577 samples, a final dataset of 114,999 EDTA plasma samples was analyzed. A detailed genetic association analysis for the investigated PUFAs is presented in Table 1.
Genetic associations with osteomyelitis
Genetic association estimates for single nucleotide polymorphisms (SNPs) linked to osteomyelitis were acquired, as indicated by IEU GWAS ID: ieu-b-4975. The dataset comprises 486,484 individuals, including 4,836 osteomyelitis cases and 481,648 controls. Data for these genetic associations were obtained from the UK Biobank references24,25 and are available in the Integrative Epidemiologic Unit (IEU) GWAS database. For a detailed overview of the genetic associations on osteomyelitis, Table 1 can be consulted. Detailed descriptions of participant recruitment and diagnostic criteria are available in the original studies. All data used in this research were obtained from publicly accessible GWAS. Since our analysis exclusively involved anonymized summary-level data already available in the public domain, ethical approval was not required for this study.
Instrumental variables selection
To delve into the potential cause-and-effect relationship between PUFAs and osteomyelitis, this study used instrumental variables (IVs) within an MR framework. These IVs served as proxies and were selected after an extensive review of GWAS data for significant SNPs associated with PUFAs. SNPs were chosen based on their strong genome-wide associations with PUFAs, meeting a significance threshold (p < 5 × 10−8). The quality and independence of these IVs were ensured by filtering for a sizeable genomic distance between SNPs (over 10,000 kb) and low linkage disequilibrium (r2 < 0.001). Palindromic SNPs, associated with the outcome at the genome-wide significance level, or absent from the outcome GWAS summary data were excluded.
To ensure a robust association between these IVs and PUFAs, we removed SNPs with F statistics less than 10. The equation for computing the F-statistic is expressed as F = ((N-2) × R2/(1-R2)), where R2 denotes the proportion of variance in the exposure explained by the SNP, and N represents the sample size26. The formula for calculating R2 is articulated as follows: R2 = 2 × MAF × (1 − MAF) × (β2/ (SE2 × N)), wherein MAF stands for the minor allele frequency, β represents the effect size of the exposure, and SE signifies the standard error of the effect size. Based on these rigorous selection criteria and the use of these IVs, we were able to establish strong and reliable causal associations between PUFAs and osteomyelitis risk.
MR analysis
In this research, three established analytical methods were used to explore the association between PUFAs and osteomyelitis, namely the inverse-variance weighted (IVW), MR-Egger, and weighted median techniques. The IVW method was the main method applied in the MR analysis due to its substantial statistical power, aggregating the Wald ratio estimates of individual SNPs to generate an integrated causal estimate. The MR-Egger method was also employed to yield consistent estimations of causative effects, factoring in the instrument strength independently from the direct effects27. Additionally, the weighted median approach was used, providing trustworthy estimates from instrumental variables, which contribute over half the weight in the analysis28. To ensure the dependability of the results, it was required that the findings from the IVW method be statistically significant, and the results from the weighted median and MR-Egger methods should be directionally aligned with IVW results.
MRlap analysis
Since our summary-level genetic data for exposure and outcome were both derived from European populations, we could not fully exclude potential sample overlap. Sample overlap can bias Mendelian Randomization (MR) estimates towards observational associations. To address this issue, we employed the MRlap (Mendelian Randomization with Laplacian correction) method, which adjusts MR estimates for bias introduced by overlapping samples. Specifically, MRlap evaluates whether differences between the original (IVW-based) MR results and overlap-corrected estimates are statistically significant. A non-significant difference (p > 0.05) between observed and MRlap-corrected effects indicates minimal bias due to sample overlap, suggesting the IVW estimates remain reliable. In contrast, a significant difference (p < 0.05) suggests substantial overlap bias, and the MRlap-adjusted estimates should be considered more accurate.
Heterogeneity, horizontal pleiotropy and sensitivity analysis
To assess heterogeneity across individual SNP estimates, the study used Cochran’s Q test, with significance determined at a p-value less than 0.05, signifying potential variability. It should be recognized that heterogeneity does not automatically discredit the inverse-variance weighted (IVW) method outcomes. The potential for horizontal pleiotropy was examined using the MR-Egger intercept test, where an intercept significantly different from zero implies pleiotropic effects. This test was instrumental in determining whether factors other than the primary exposure might be influencing the results. A variety of sensitivity analyses were conducted to confirm the integrity of the study’s conclusions. For visual assessment of the relationship between PUFAs and osteomyelitis, scatter plots were examined. These plots visually confirmed the direction and consistency of associations between PUFAs and osteomyelitis. The Leave-One-Out analysis technique was implemented, removing SNPs one at a time showed that no single SNP disproportionately influenced the results, supporting robustness. Funnel plots were also reviewed to detect any asymmetry that could suggest horizontal pleiotropy. The symmetry observed in funnel plots suggested an absence of significant bias, reinforcing the validity of the findings. These analyses were vital in reinforcing the Mendelian randomization findings’ credibility. We further conducted MR-PRESSO (Mendelian Randomization Pleiotropy RESidual Sum and Outlier) analysis to detect and correct for potential horizontal pleiotropy by identifying significant outlier SNPs (p < 0.05).
To account for multiple testing, we applied a Bonferroni correction based on the number of exposures analyzed (five PUFAs), setting the significance threshold at P < 0.01 (0.05/5). Associations with P-values between 0.01 and 0.05 were considered suggestive and interpreted with caution. Calculations of the odds ratios (ORs) for the effect of PUFAs on osteomyelitis risk, along with their 95% confidence intervals, were conducted. Statistical computations were performed using the R software environment, particularly employing the “TwoSampleMR” and “MRlap” package, which is designed to provide robust and controlled evaluations of causal relationships.
Results
Characteristics of genetic variants
The genome-wide significance threshold of 5 × 10−8 enabled the identification of multiple SNPs associated with various polyunsaturated fatty acids (PUFAs); 35 SNPs were linked to omega-3, 43 to omega-6, 27 to DHA, and 37 to LA. SNPs associated with the omega-6 to omega-3 ratio numbered at 30. Notably, all F statistic values for these SNPs were above 10, suggesting a strong linkage between the instrumental variables (IVs) and the PUFAs in question. For a detailed account, Supplementary Tables S1–S5 contained these specifics.
MR analysis: PUFAs as exposure, osteomyelitis as outcome
The MR analysis, with PUFAs as the exposure and osteomyelitis as the outcome, is presented in Fig. 2. The IVW method provided preliminary indications of a suggestive positive risk of osteomyelitis linked to genetically predicted DHA (OR 1.158, 95% CI 1.030–1.301, P = 0.014) and omega 6 (OR 1.156, 95% CI 1.009–1.325, P = 0.037). Moreover, a suggestive reduced risk associated with the omega-6 to omega-3 ratio (OR 0.905, 95% CI 0.821–0.999, P = 0.048). No significant associations were observed for LA (OR 1.046, 95% CI 0.912–1.199, P = 0.522) or omega-3 (OR 1.082, 95% CI 0.983–1.192, P = 0.107) with osteomyelitis. The MR-Egger, weighted median, and weighted mode analyses corroborated the IVW method results, except in the case of omega-6. While the IVW method indicated significant associations, the MR-Egger and weighted median methods showed wider confidence intervals, suggesting some uncertainty.
The forest plot of IVW, MR-Egger and weighted median. The error bars represent the 95% CIs of the MR estimates in OR per SD change in genetically determined liver and pancreas fat/volume. MR, mendelian randomization; IVW, Inverse variance weighted; DHA, docosahexaenoic acid; LA, linoleic acid; omega-3, omega-3 fatty acids; omega-6, omega-6 fatty acids; omega-6:3, the omega-6 to omega-3 fatty acid ratio.
MRlap analysis
The MRlap-corrected results shown in Table 3 were consistent with those obtained from the primary MR analyses, confirming the robustness of the original IVW method estimates and indicating minimal bias due to potential sample overlap.
Heterogeneity, horizontal pleiotropy, and sensitivity analysis
Table 2 details the outcomes from Cochran’s Q test and the test for pleiotropy. The MR analyses did not reveal any notable heterogeneity, and the MR-Egger intercept tests indicated an absence of horizontal pleiotropy across the analyses, with all p-values exceeding 0.05. Scatter plots and forest plots, which provide further insight, can be found in the supplementary materials (Supplementary Figs. 1 and 2). The leave-one-out plot demonstrates that there was potentially influential SNPs driving the causal link for LA and omega-3 on osteomyelitis, and the current findings need to be interpreted carefully with caution (Supplementary Fig. 3). Additionally, the symmetry observed in the funnel plots for all MR analyses suggests the non-existence of horizontal pleiotropy, with the variation in effect size around the point estimate appearing symmetrical (Supplementary Fig. 4). The MR-PRESSO analysis similarly found no evidence of significant horizontal pleiotropy or outlier SNPs (all p-values > 0.05), further supporting the robustness of our results (Table 3).
Discussion
Our MR analysis provides suggestive evidence for associations between specific PUFAs and osteomyelitis risk. Genetically predicted levels of DHA and omega-6 fatty acids were associated with a suggestive increase in osteomyelitis risk, whereas a higher omega-6 to omega-3 ratio was associated with a suggestive reduction in risk. Although these findings do not meet the threshold for definitive conclusions, they highlight intriguing trends that merit further exploration.
In contrast to recently published experimental studies16,29, our Mendelian randomization analysis did not confirm the inverse association between increasing omega-3 intake, either through supplementation or diet, and bacterial growth and bone infection. Instead, our findings provided weak evidence that a genetically predicted higher DHA is associated with a higher risk of osteomyelitis. While these results may seem counterintuitive due to the anti-inflammatory properties of DHA30, several factors should be considered to shed light on these associations. First, the study used non-fasting plasma samples for PUFA testing, which can introduce variability in fatty acid concentrations, as they are influenced by recent dietary intake. DHA is traditionally recognized for its anti-inflammatory properties, particularly through its role in producing pro-resolving lipid mediators such as resolvins, which help to terminate inflammation. However, in the context of chronic infections like osteomyelitis, DHA immunomodulatory effects may shift towards an immune tolerance profile that inadvertently supports pathogen persistence rather than clearance. A recent study indicate that osteomyelitis is characterized by an immune environment rich in immune exhaustion markers (e.g., PD-1, TIM-3, VISTA) and regulatory immune cells (e.g., Tregs and Bregs) that facilitate immune evasion by pathogens such as Staphylococcus aureus31. This represents a microbial strategy to attenuate the host immune response by upregulating the expression of exhaustion markers on immune cells and shifting the immune system towards an anti-inflammatory phenotype32. In this environment, the role of DNA in promoting anti-inflammatory pathways may suppress effective immune responses, potentially aiding in the establishment and maintenance of chronic infection. We propose that further studies are warranted to explore the dual role of DHA in inflammation, especially distinguishing its impact on immune responses in acute versus chronic infection contexts. Individual genetic differences can significantly impact how the body metabolizes and responds to specific nutrients33. It is possible that genetic factors play a role in the observed associations. Some individuals may possess genetic variations that modify their responses to certain PUFAs, influencing their susceptibility to osteomyelitis34,35.
In this study, no statistically significant difference was found for genetically predicted linoleic acid, omega-3 levels, and omega-6 fatty acids with the risk of osteomyelitis, but an association was found with the omega-6 to omega-3 ratio. This finding aligns with studies on other inflammatory diseases, such as rheumatoid arthritis and cardiovascular disease, which have also indicated that the balance between omega-6 and omega-3 fatty acids is crucial for modulating immune response and inflammation8,36. This highlights the complexity of understanding the impact of omega-3 and omega-6 intake on osteomyelitis risk compared to other infection diseases and underscores the importance of considering evidence from multiple sources when making conclusions. The production of prostaglandin E2 (PGE2) in bone was significantly lower in animals that were fed diets with a lower dietary ratio of omega-6 to omega-3 PUFAs compared to those rats that were fed diets with a higher dietary ratio37,38. This reduction in PGE2 has been associated with a lower osteoclast activity and reduced bone resorption, contributing to a more favorable condition for bone integrity and highlighting the potential role of omega-3 in offsetting omega-6-driven inflammation in bone health. In osteomyelitis, the infection can lead to the destruction of bone tissue and PGE2 may contribute to this bone loss by stimulating the activity of cells called osteoclasts, which are responsible for breaking down bone39. The observed changes in PGE2 levels reflect a more favorable condition for bone formation and reduced bone resorption. However, further research and clinical studies are needed to confirm these effects and develop comprehensive strategies for osteomyelitis management. The interplay between dietary factors and bone health is complex, and this study provides a valuable starting point for future investigations.
Moreover, genetic variations that influence DHA metabolism and signaling could modulate individual susceptibility to such effects. Some individuals may carry genetic variants that promote a stronger immune-regulatory response to DHA, which could elevate their risk of chronic infections under certain conditions. This aligns with emerging evidence on gene-diet interactions, suggesting that genetic predispositions may critically shape the immunomodulatory role of specific PUFAs like DHA. These findings underscore the importance of considering both genetic and environmental factors when interpreting the physiological actions of DHA in infectious diseases like osteomyelitis.
The study is limited by the study focusing on individuals of European ancestry, which may limit the generalizability of the findings to other populations. Second, genetic and environmental factors related to osteomyelitis can vary among different ethnic groups, and caution should be exercised when applying these findings to a more diverse population. The study relies on large-scale GWAS summary data, which might not capture all relevant genetic and environmental factors influencing osteomyelitis risk. Individual dietary habits, physical activity, and other lifestyle factors could confound the observed associations. Further research is needed to translate these associations into actionable strategies for osteomyelitis prevention and treatment. It should focus on longitudinal studies and experimental models to explore causality and mechanisms.
In summary, our MR analysis suggests potential associations between elevated genetically predicted levels of DHA and omega-6 fatty acids with osteomyelitis risk, and a possible protective effect of a higher omega-6 to omega-3 ratio. While these associations are suggestive and should be interpreted cautiously, they underscore the need for further investigation into the complex role of PUFAs in chronic bone infections. Future research could explore the genetic and immune interactions that influence PUFA-related effects on osteomyelitis, potentially guiding personalized dietary or therapeutic strategies aimed at reducing osteomyelitis risk and improving outcomes for affected individuals.
Data availability
All the Mendelian randomization study files are available from GWAS (https://gwas.mrcieu.ac.uk ). GWAS ID: ieu-b-4975 (https://gwas.mrcieu.ac.uk/datasets/ieu-b-4975/), met-d-Omega_3 (https://gwas.mrcieu.ac.uk/datasets/ met-d-Omega_3/), met-d-Omega_6 (https://gwas.mrcieu.ac.uk/datasets/ met-d-Omega_6/), met-d-Omega_6_by_Omega_3 (https://gwas.mrcieu.ac.uk/datasets/met-d-Omega_6_by_Omega_3/), met-d-DHA (https://gwas.mrcieu.ac.uk/datasets/ met-d-DHA /), met-d-LA (https://gwas.mrcieu.ac.uk/datasets/ met-d-LA /)). Researchers seeking additional details regarding the datasets used in this study may contact Hang Dong.
References
Moriarty, T. F. et al. Bone infection: A clinical priority for clinicians, scientists and educators. Eur. Cell Mater. 42, 312–333. https://doi.org/10.22203/eCM.v042a21 (2021).
Masters, E. A. et al. Skeletal infections: Microbial pathogenesis, immunity and clinical management. Nat. Rev. Microbiol. 20, 385–400. https://doi.org/10.1038/s41579-022-00686-0 (2022).
Hofstee, M. I. et al. Current concepts of osteomyelitis: From pathologic mechanisms to advanced research methods. Am. J. Pathol. 190, 1151–1163. https://doi.org/10.1016/j.ajpath.2020.02.007 (2020).
Koehler, P., Tacke, D. & Cornely, O. A. Aspergillosis of bones and joints—A review from 2002 until today. Mycoses 57, 323–335. https://doi.org/10.1111/myc.12165 (2014).
Lew, D. P. & Waldvogel, F. A. Osteomyelitis. Lancet 364, 369–379. https://doi.org/10.1016/S0140-6736(04)16727-5 (2004).
Xie, X. et al. Genetic determinants for bacterial osteomyelitis: A focused systematic review of published literature. Front. Genet. 12, 654792. https://doi.org/10.3389/fgene.2021.654792 (2021).
Garcia Del Pozo, E., Collazos, J., Carton, J. A., Camporro, D. & Asensi, V. Factors predictive of relapse in adult bacterial osteomyelitis of long bones. BMC Infect. Dis. 18, 635. https://doi.org/10.1186/s12879-018-3550-6 (2018).
Djuricic, I. & Calder, P. C. Beneficial outcomes of Omega-6 and Omega-3 polyunsaturated fatty acids on human health: An update for 2021. Nutrients https://doi.org/10.3390/nu13072421 (2021).
Gutierrez, S., Svahn, S. L. & Johansson, M. E. Effects of Omega-3 fatty acids on immune cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20205028 (2019).
Molfino, A., Amabile, M. I., Monti, M. & Muscaritoli, M. Omega-3 polyunsaturated fatty acids in critical illness: Anti-inflammatory, proresolving, or both?. Oxid. Med. Cell Longev. 2017, 5987082. https://doi.org/10.1155/2017/5987082 (2017).
Ferguson, J. F. et al. Omega-3 PUFA supplementation and the response to evoked endotoxemia in healthy volunteers. Mol. Nutr. Food Res. 58, 601–613. https://doi.org/10.1002/mnfr.201300368 (2014).
Chen, Y. L. et al. Omega-3 fatty acids impair miR-1-3p-dependent Notch3 down-regulation and alleviate sepsis-induced intestinal injury. Mol. Med. 28, 9. https://doi.org/10.1186/s10020-021-00425-w (2022).
Korner, A. et al. Resolution of inflammation and sepsis survival are improved by dietary Omega-3 fatty acids. Cell Death Differ. 25, 421–431. https://doi.org/10.1038/cdd.2017.177 (2018).
Wolbrink, D. R. J. et al. Are omega-3 fatty acids safe and effective in acute pancreatitis or sepsis? A systematic review and meta-analysis. Clin. Nutr. 39, 2686–2694. https://doi.org/10.1016/j.clnu.2019.12.006 (2020).
Boivin, A. et al. Docosahexaenoic acid, but not eicosapentaenoic acid, improves septic shock-induced arterial dysfunction in rats. PLoS ONE 12, e0189658. https://doi.org/10.1371/journal.pone.0189658 (2017).
Zhou, P. et al. The synergistic therapeutic efficacy of vancomycin and omega-3 fatty acids alleviates Staphylococcus aureus-induced osteomyelitis in rats. Biomed. Pharmacother. 111, 1228–1233. https://doi.org/10.1016/j.biopha.2018.12.125 (2019).
Burgess, S. et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 4, 186. https://doi.org/10.12688/wellcomeopenres.15555.3 (2019).
Smith, G. D. & Ebrahim, S. Mendelian randomization: Prospects, potentials, and limitations. Int. J. Epidemiol. 33, 30–42. https://doi.org/10.1093/ije/dyh132 (2004).
Burgess, S., Daniel, R. M., Butterworth, A. S. & Thompson, S. G. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int. J. Epidemiol. 44, 484–495. https://doi.org/10.1093/ije/dyu176(2015) (2015).
Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: The STROBE-MR statement. JAMA 326, 1614–1621. https://doi.org/10.1001/jama.2021.18236 (2021).
Shin, S. Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550. https://doi.org/10.1038/ng.2982 (2014).
Li, Y., Li, Q., Cao, Z. & Wu, J. The causal association of polyunsaturated fatty acids with allergic disease: A two-sample Mendelian randomization study. Front. Nutr. 9, 962787. https://doi.org/10.3389/fnut.2022.962787 (2022).
Julkunen, H., Cichonska, A., Slagboom, P. E., Wurtz, P. & Nightingale Health, U. K. B. I. Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. Elife https://doi.org/10.7554/eLife.63033 (2021).
Chen, B., Pu, B., Li, S., Gong, Y. & Dong, H. The role of NSAID in mediating the effect of genetically predicted major depressive disorder on osteomyelitis: A Mendelian randomization study. J. Affect. Disord. 341, 62–66. https://doi.org/10.1016/j.jad.2023.08.121 (2023).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779. https://doi.org/10.1371/journal.pmed.1001779 (2015).
Burgess, S., Thompson, S. G. & Collaboration, C. C. G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764. https://doi.org/10.1093/ije/dyr036 (2011).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. https://doi.org/10.1093/ije/dyv080 (2015).
Bowden, J., DaveySmith, G., Haycock, P. C. & Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 40, 304–314. https://doi.org/10.1002/gepi.21965 (2016).
Coraca-Huber, D. C., Steixner, S., Wurm, A. & Nogler, M. Antibacterial and anti-biofilm activity of Omega-3 polyunsaturated fatty acids against periprosthetic joint infections-isolated multi-drug resistant strains. Biomedicines. https://doi.org/10.3390/biomedicines9040334 (2021).
Kuda, O. et al. Docosahexaenoic acid-derived fatty acid esters of hydroxy fatty acids (FAHFAs) with anti-inflammatory properties. Diabetes 65, 2580–2590. https://doi.org/10.2337/db16-0385 (2016).
Surendar, J. et al. Osteomyelitis is associated with increased anti-inflammatory response and immune exhaustion. Front. Immunol. 15, 1396592. https://doi.org/10.3389/fimmu.2024.1396592 (2024).
Rogovskii, V. Immune tolerance as the physiologic counterpart of chronic inflammation. Front. Immunol. 11, 2061. https://doi.org/10.3389/fimmu.2020.02061 (2020).
Elsamanoudy, A. Z., Mohamed Neamat-Allah, M. A., Hisham Mohammad, F. A., Hassanien, M. & Nada, H. A. The role of nutrition related genes and nutrigenetics in understanding the pathogenesis of cancer. J. Microsc. Ultrastruct. 4, 115–122. https://doi.org/10.1016/j.jmau.2016.02.002 (2016).
Mathias, R. A., Pani, V. & Chilton, F. H. Genetic variants in the FADS gene: Implications for dietary recommendations for fatty acid intake. Curr. Nutr. Rep. 3, 139–148. https://doi.org/10.1007/s13668-014-0079-1 (2014).
Glaser, C., Lattka, E., Rzehak, P., Steer, C. & Koletzko, B. Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern. Child Nutr. 7(Suppl 2), 27–40. https://doi.org/10.1111/j.1740-8709.2011.00319.x (2011).
Yates, C. M., Calder, P. C. & Ed Rainger, G. Pharmacology and therapeutics of omega-3 polyunsaturated fatty acids in chronic inflammatory disease. Pharmacol. Ther. 141, 272–282. https://doi.org/10.1016/j.pharmthera.2013.10.010 (2014).
Watkins, B. A., Li, Y., Allen, K. G., Hoffmann, W. E. & Seifert, M. F. Dietary ratio of (n-6)/(n-3) polyunsaturated fatty acids alters the fatty acid composition of bone compartments and biomarkers of bone formation in rats. J. Nutr. 130, 2274–2284. https://doi.org/10.1093/jn/130.9.2274 (2000).
Al-Nouri, D., Al-Khalifa, A. & Shahidi, F. Long-term supplementation of dietary omega-6/omega-3 ratios alters bone marrow fatty acid and biomarkers of bone metabolism in growing rabbits. J. Funct. Foods 4, 584–593 (2012).
Kelly, O., Cusack, S., Jewell, C. & Cashman, K. D. The effect of polyunsaturated fatty acids, including conjugated linoleic acid, on calcium absorption and bone metabolism and composition in young growing rats. Br. J. Nutr. 90, 743–750. https://doi.org/10.1079/bjn2003951 (2003).
Acknowledgements
The authors are thankful for all the funders, participants, and investigators of the UK Biobank
Funding
This work was funded by National Natural Science Foundation of China (82004390), Guangzhou Municipal School-Enterprise Joint Funding Project (2025A03J3892), National Studio Construction projects for the famous experts in Traditional Chinese Medicine (Huang Feng studio N75, 2022) and China Scholarship Council.
Author information
Authors and Affiliations
Contributions
BC: study conception and design, acquisition of data, analysis and interpretation of data, writing manuscript; BP: critical revision of manuscript, analysis and interpretation of data; SL: acquisition of data, statistics analysis; SSL: acquisition of data, critical revision of manuscript; HD: study conception and design, drafting of manuscript, critical revision of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was conducted using publicly available summary statistics from genome-wide association studies (GWAS) and data from the UK Biobank. All methods were carried out in accordance with relevant guidelines and regulations. The UK Biobank has obtained ethics approval from the Northwest Multi-Centre Research Ethics Committee (11/NW/0382), and all participants provided informed consent at the time of enrollment. No new individual-level data collection was performed for this study, and ethical approval was not required for the use of publicly available summary-level data.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, B., Pu, B., Lin, S. et al. Investigating the association between polyunsaturated fatty acids and osteomyelitis by Mendelian randomization. Sci Rep 15, 14760 (2025). https://doi.org/10.1038/s41598-025-98502-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-98502-1